Tag-based next generation sequencing: a feasible and reliable assay for EGFR T790M mutation detection in circulating tumor DNA of non small cell lung cancer patients
- PMID: 31029076
- PMCID: PMC6487061
- DOI: 10.1186/s10020-019-0082-5
Tag-based next generation sequencing: a feasible and reliable assay for EGFR T790M mutation detection in circulating tumor DNA of non small cell lung cancer patients
Abstract
Background: The demonstration of EGFR T790M gene mutation in plasma is crucial to assess the eligibility of Non Small Cell Lung Cancer (NSCLC) patients, who have acquired resistance to first or second generation Tyrosine Kinase Inhibitors (TKIs), to receive a subsequent treatment with osimertinib. Since circulating tumor DNA (ctDNA) is present in very low amounts in plasma, high sensitive and specific methods are required for molecular analysis. Improving sensitivity of T790M mutation detection in plasma ctDNA enables a larger number of NSCLC patients to receive the appropriate therapy without any further invasive procedure.
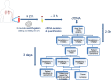
Methods: A tag-based next generation sequencing (NGS) platform capable of tagging rare circulating tumor DNA alleles was employed in this study for the identification of T790M mutation in 42 post-TKI NSCLC patients.
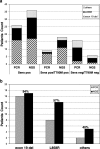
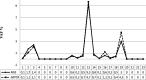
Results: Compared to Real Time PCR, tag-based NGS improved the T790M detection rate (42.85% versus 21.4%, respectively), especially in those cases with a low median mutation abundance (i.e. 0.24, range 0.07-0.78). Moreover, the tag-based NGS identified EGFR activating mutations more efficiently than Real Time PCR (85.7% versus 61.9% detection rate, respectively), particularly of the L858R variant type (0.06-0.75 mutation abundance range). Patients in whom the T790M mutation was detected in plasma, achieved an objective response to osimertinib (9/14, 64.28%).
Conclusions: Tag-based NGS represents an accurate and sensitive tool in a clinical setting for non-invasive assessment and monitoring of T790M variant in NSCLC patients.
Keywords: C797S; Circulating tumor DNA; EGFR TKIs; Liquid biopsy; Molecular tag; NSCLC; Next generation sequencing; T790M resistance mutation.
Conflict of interest statement
Ethics approval and consent to participate
All of the procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions. Written informed consent was obtained from each patient included in the study. The study protocol has been approved by the Ethics Committee of Liguria Region (Italy) (P.R.273REG2016).
Consent for publication
Not applicable.
Competing interests
Drs. M. Dono and S. Zupo received speaker honoraria from AstraZeneca; Dr. C. Genova discloses being advisory Board member of AstraZeneca and received honoraria from AstraZeneca, Boehringer-Ingelheim, Bristol-Myers-Squibb, Merck Sharp & Dohme and Roche; Dr. F. Grossi received honoraria from AMGEN, AstraZeneca, Bristol-Myers Squibb, Boehringer Ingelheim, Celgene, Merck Sharp & Dohme, Pfizer, Pierre Fabre, Roche.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
High MAF of EGFR mutations and high ratio of T790M sensitizing mutations in ctDNA predict better third-generation TKI outcomes.Thorac Cancer. 2020 Jun;11(6):1503-1511. doi: 10.1111/1759-7714.13418. Epub 2020 Apr 14. Thorac Cancer. 2020. PMID: 32285618 Free PMC article.
-
Clinical utility of plasma-based digital next-generation sequencing in oncogene-driven non-small-cell lung cancer patients with tyrosine kinase inhibitor resistance.Lung Cancer. 2019 Aug;134:72-78. doi: 10.1016/j.lungcan.2019.05.032. Epub 2019 May 30. Lung Cancer. 2019. PMID: 31319999
-
[Detection of circulating tumor DNA in epidermal growth factor receptor-TKI relapsed non-small cell lung cancer patients using next-generation sequencing and an analysis of the resistant mechanisms].Zhonghua Bing Li Xue Za Zhi. 2018 Dec 8;47(12):904-909. doi: 10.3760/cma.j.issn.0529-5807.2018.12.002. Zhonghua Bing Li Xue Za Zhi. 2018. PMID: 30522169 Chinese.
-
Targeting EGFRL858R/T790M and EGFRL858R/T790M/C797S resistance mutations in NSCLC: Current developments in medicinal chemistry.Med Res Rev. 2018 Sep;38(5):1550-1581. doi: 10.1002/med.21488. Epub 2018 Jan 26. Med Res Rev. 2018. PMID: 29377179 Review.
-
Evidence-based best practices for EGFR T790M testing in lung cancer in Canada.Curr Oncol. 2018 Apr;25(2):163-169. doi: 10.3747/co.25.4044. Epub 2018 Apr 30. Curr Oncol. 2018. PMID: 29719432 Free PMC article.
Cited by
-
The Role of Circulating Biomarkers in Lung Cancer.Front Oncol. 2022 Jan 21;11:801269. doi: 10.3389/fonc.2021.801269. eCollection 2021. Front Oncol. 2022. PMID: 35127511 Free PMC article. Review.
-
Liquid Biopsy in Non-Small Cell Lung Cancer: Highlights and Challenges.Cancers (Basel). 2019 Dec 19;12(1):17. doi: 10.3390/cancers12010017. Cancers (Basel). 2019. PMID: 31861557 Free PMC article. Review.
-
Optimization of a WGA-Free Molecular Tagging-Based NGS Protocol for CTCs Mutational Profiling.Int J Mol Sci. 2020 Jun 19;21(12):4364. doi: 10.3390/ijms21124364. Int J Mol Sci. 2020. PMID: 32575430 Free PMC article.
-
Comparison of liquid-based to tissue-based biopsy analysis by targeted next generation sequencing in advanced non-small cell lung cancer: a comprehensive systematic review.J Cancer Res Clin Oncol. 2020 Aug;146(8):2051-2066. doi: 10.1007/s00432-020-03267-x. Epub 2020 May 27. J Cancer Res Clin Oncol. 2020. PMID: 32462295 Free PMC article.
-
Clinical feasibility of NGS liquid biopsy analysis in NSCLC patients.PLoS One. 2019 Dec 20;14(12):e0226853. doi: 10.1371/journal.pone.0226853. eCollection 2019. PLoS One. 2019. PMID: 31860648 Free PMC article.
References
-
- Ariyasu R, Nishikawa S, Uchibori K, et al. High ratio of T790M to EGFR activating mutations correlate with the osimertinib response in non-small-cell lung cancer. Lung Cancer. 2018;117:1–6. - PubMed
-
- Arulananda S, Do H, Musafer A, Mitchell P, Dobrovic A, John T. Combination Osimertinib and Gefitinib in C797S and T790M EGFR-mutated non-small cell lung Cancer. J Thorac Oncol. 2017;12(11):1728–1732. - PubMed
-
- Bartels S, Persing S, Hasemeier B, Schipper E, Kreipe H, Lehmann H. Molecular analysis of circulating cell-free DNA from lung Cancer patients in routine laboratory practice. A Cross-Platform Comparison of Three Different Molecular Methods for Mutation Detection. J Mol Diagn. 2017;19:722. - PubMed
-
- Coco S, Truini A, Vanni I, et al. Next generation sequencing in non-small cell lung cancer: new avenues toward the personalized medicine. Curr Drug Targets. 2015;16(1):47–59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

