Is population structure in the genetic biobank era irrelevant, a challenge, or an opportunity?
- PMID: 31030318
- PMCID: PMC6942007
- DOI: 10.1007/s00439-019-02014-8
Is population structure in the genetic biobank era irrelevant, a challenge, or an opportunity?
Erratum in
-
Correction to: Is population structure in the genetic biobank era irrelevant, a challenge, or an opportunity?Hum Genet. 2020 Jan;139(1):43-44. doi: 10.1007/s00439-019-02051-3. Hum Genet. 2020. PMID: 31363835 Free PMC article.
Abstract
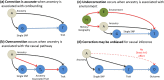
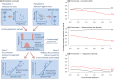
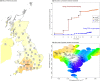
Replicable genetic association signals have consistently been found through genome-wide association studies in recent years. The recent dramatic expansion of study sizes improves power of estimation of effect sizes, genomic prediction, causal inference, and polygenic selection, but it simultaneously increases susceptibility of these methods to bias due to subtle population structure. Standard methods using genetic principal components to correct for structure might not always be appropriate and we use a simulation study to illustrate when correction might be ineffective for avoiding biases. New methods such as trans-ethnic modeling and chromosome painting allow for a richer understanding of the relationship between traits and population structure. We illustrate the arguments using real examples (stroke and educational attainment) and provide a more nuanced understanding of population structure, which is set to be revisited as a critical aspect of future analyses in genetic epidemiology. We also make simple recommendations for how problems can be avoided in the future. Our results have particular importance for the implementation of GWAS meta-analysis, for prediction of traits, and for causal inference.
Conflict of interest statement
DJL is a director of GENSCI Ltd. On behalf of all authors, the corresponding author states that there is no other conflict of interest.
Figures


References
-
- Adhikari K, Fontanil T, Cal S, Mendoza-Revilla J, Fuentes-Guajardo M, Chacón-Duque J-C, Al-Saadi F, Johansson JA, Quinto-Sanchez M, Acuña-Alonzo V, Jaramillo C, Arias W, Lozano RB, Pérez GM, Gómez-Valdés J, Villamil-Ramírez H, Hunemeier T, Ramallo V, de Cerqueira CCS, Hurtado M, Villegas V, Granja V, Gallo C, Poletti G, Schuler-Faccini L, Salzano FM, Bortolini M-C, Canizales-Quinteros S, Rothhammer F, Bedoya G, Gonzalez-José R, Headon D, López-Otín C, Tobin DJ, Balding D, Ruiz-Linares A. A genome-wide association scan in admixed Latin Americans identifies loci influencing facial and scalp hair features. Nat Commun. 2016;7:10815. doi: 10.1038/ncomms10815. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

