Force-Regulated Refolding of the Mechanosensory Domain in the Platelet Glycoprotein Ib-IX Complex
- PMID: 31030883
- PMCID: PMC6531785
- DOI: 10.1016/j.bpj.2019.03.037
Force-Regulated Refolding of the Mechanosensory Domain in the Platelet Glycoprotein Ib-IX Complex
Abstract
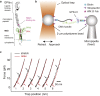
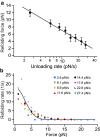
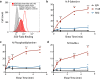
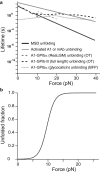
In platelets, the glycoprotein (GP) Ib-IX receptor complex senses blood shear flow and transmits the mechanical signals into platelets. Recently, we have discovered a juxtamembrane mechanosensory domain (MSD) within the GPIbα subunit of GPIb-IX. Mechanical unfolding of the MSD activates GPIb-IX signaling into platelets, leading to their activation and clearance. Using optical tweezer-based single-molecule force measurement, we herein report a systematic biomechanical characterization of the MSD in its native, full-length receptor complex and a recombinant, unglycosylated MSD in isolation. The native MSD unfolds at a resting rate of 9 × 10-3 s-1. Upon exposure to pulling forces, MSD unfolding accelerates exponentially over a force scale of 2.0 pN. Importantly, the unfolded MSD can refold with or without applied forces. The unstressed refolding rate of MSD is ∼17 s-1 and slows exponentially over a force scale of 3.7 pN. Our measurements confirm that the MSD is relatively unstable, with a folding free energy of 7.5 kBT. Because MSD refolding may turn off GPIb-IX's mechanosensory signals, our results provide a mechanism for the requirement of a continuous pulling force of >15 pN to fully activate GPIb-IX.
Copyright © 2019 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Peterson D.M., Stathopoulos N.A., Moake J.L. Shear-induced platelet aggregation requires von Willebrand factor and platelet membrane glycoproteins Ib and IIb-IIIa. Blood. 1987;69:625–628. - PubMed
-
- Savage B., Saldívar E., Ruggeri Z.M. Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell. 1996;84:289–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

