Marine DNA Viral Macro- and Microdiversity from Pole to Pole
- PMID: 31031001
- PMCID: PMC6525058
- DOI: 10.1016/j.cell.2019.03.040
Marine DNA Viral Macro- and Microdiversity from Pole to Pole
Abstract
Microbes drive most ecosystems and are modulated by viruses that impact their lifespan, gene flow, and metabolic outputs. However, ecosystem-level impacts of viral community diversity remain difficult to assess due to classification issues and few reference genomes. Here, we establish an ∼12-fold expanded global ocean DNA virome dataset of 195,728 viral populations, now including the Arctic Ocean, and validate that these populations form discrete genotypic clusters. Meta-community analyses revealed five ecological zones throughout the global ocean, including two distinct Arctic regions. Across the zones, local and global patterns and drivers in viral community diversity were established for both macrodiversity (inter-population diversity) and microdiversity (intra-population genetic variation). These patterns sometimes, but not always, paralleled those from macro-organisms and revealed temperate and tropical surface waters and the Arctic as biodiversity hotspots and mechanistic hypotheses to explain them. Such further understanding of ocean viruses is critical for broader inclusion in ecosystem models.
Keywords: community ecology; diversity gradients; marine biology; metagenomics; population ecology; species; viruses.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
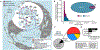


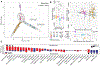

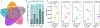

Comment in
-
Drowning in Viruses.Cell. 2019 May 16;177(5):1084-1085. doi: 10.1016/j.cell.2019.04.045. Cell. 2019. PMID: 31100262
References
-
- Achtman M, and Wagner M (2008). Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol 6, 431–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

