Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia
- PMID: 31031138
- PMCID: PMC6617387
- DOI: 10.1016/j.stem.2019.03.021
Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia
Abstract
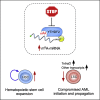
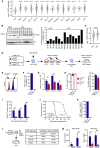
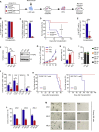
Acute myeloid leukemia (AML) is an aggressive clonal disorder of hematopoietic stem cells (HSCs) and primitive progenitors that blocks their myeloid differentiation, generating self-renewing leukemic stem cells (LSCs). Here, we show that the mRNA m6A reader YTHDF2 is overexpressed in a broad spectrum of human AML and is required for disease initiation as well as propagation in mouse and human AML. YTHDF2 decreases the half-life of diverse m6A transcripts that contribute to the overall integrity of LSC function, including the tumor necrosis factor receptor Tnfrsf2, whose upregulation in Ythdf2-deficient LSCs primes cells for apoptosis. Intriguingly, YTHDF2 is not essential for normal HSC function, with YTHDF2 deficiency actually enhancing HSC activity. Thus, we identify YTHDF2 as a unique therapeutic target whose inhibition selectively targets LSCs while promoting HSC expansion.
Keywords: TNFR2; YTHDF2; acute myeloid leukemia; hematopoiesis; hematopoietic stem cell; leukemic stem cells; m(6)A modification; mRNA decay.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300.
-
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- C29967/A26787/CRUK_/Cancer Research UK/United Kingdom
- 29389/CRUK_/Cancer Research UK/United Kingdom
- C29967/A14633/CRUK_/Cancer Research UK/United Kingdom
- MR/P010008/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MR/P010008/2/MRC_/Medical Research Council/United Kingdom
- 092076/WT_/Wellcome Trust/United Kingdom
- 19280/CRUK_/Cancer Research UK/United Kingdom
- 26787/CRUK_/Cancer Research UK/United Kingdom
- 106144/WT_/Wellcome Trust/United Kingdom
- 14633/CRUK_/Cancer Research UK/United Kingdom
- C5759/A20971/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

