RNA Profiling of the Human and Mouse Spinal Cord Stem Cell Niches Reveals an Embryonic-like Regionalization with MSX1+ Roof-Plate-Derived Cells
- PMID: 31031189
- PMCID: PMC6524006
- DOI: 10.1016/j.stemcr.2019.04.001
RNA Profiling of the Human and Mouse Spinal Cord Stem Cell Niches Reveals an Embryonic-like Regionalization with MSX1+ Roof-Plate-Derived Cells
Abstract
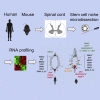
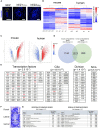
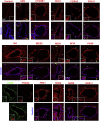
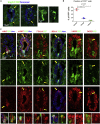
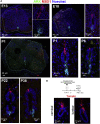
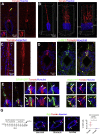
Anamniotes, rodents, and young humans maintain neural stem cells in the ependymal zone (EZ) around the central canal of the spinal cord, representing a possible endogenous source for repair in mammalian lesions. Cell diversity and genes specific for this region are ill defined. A cellular and molecular resource is provided here for the mouse and human EZ based on RNA profiling, immunostaining, and fluorescent transgenic mice. This uncovered the conserved expression of 1,200 genes including 120 transcription factors. Unexpectedly the EZ maintains an embryonic-like dorsal-ventral pattern of expression of spinal cord developmental transcription factors (ARX, FOXA2, MSX1, and PAX6). In mice, dorsal and ventral EZ cells express Vegfr3 and are derived from the embryonic roof and floor plates. The dorsal EZ expresses a high level of Bmp6 and Gdf10 genes and harbors a subpopulation of radial quiescent cells expressing MSX1 and ID4 transcription factors.
Keywords: Msx1; ependyma; ependymal cells; floor plate; neural stem cells; niche; radial glial cells; regionalization; roof plate; spinal cord; transcription factors.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Adrian E.K., Jr., Walker B.E. Incorporation of thymidine-H3 by cells in normal and injured mouse spinal cord. J. Neuropathol. Exp. Neurol. 1962;21:597–609. - PubMed
-
- Barnabé-Heider F., Göritz C., Sabelström H., Takebayashi H., Pfrieger F.W., Meletis K., Frisén J. Origin of new glial cells in intact and injured adult spinal cord. Cell Stem Cell. 2010;7:470–482. - PubMed
-
- Bauchet L., Lonjon N., Vachiery-Lahaye F., Boularan A., Privat A., Hugnot J.-P. Isolation and culture of precursor cells from the adult human spinal cord. Methods Mol. Biol. 2013;1059:87–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

