Proteomic and Isotopic Response of Desulfovibrio vulgaris to DsrC Perturbation
- PMID: 31031715
- PMCID: PMC6470260
- DOI: 10.3389/fmicb.2019.00658
Proteomic and Isotopic Response of Desulfovibrio vulgaris to DsrC Perturbation
Abstract
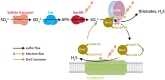
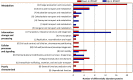
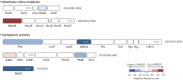
Dissimilatory sulfate reduction is a microbial energy metabolism that can produce sulfur isotopic fractionations over a large range in magnitude. Calibrating sulfur isotopic fractionation in laboratory experiments allows for better interpretations of sulfur isotopes in modern sediments and ancient sedimentary rocks. The proteins involved in sulfate reduction are expressed in response to environmental conditions, and are collectively responsible for the net isotopic fractionation between sulfate and sulfide. We examined the role of DsrC, a key component of the sulfate reduction pathway, by comparing wildtype Desulfovibrio vulgaris DSM 644T to strain IPFG07, a mutant deficient in DsrC production. Both strains were cultivated in parallel chemostat reactors at identical turnover times and cell specific sulfate reduction rates. Under these conditions, sulfur isotopic fractionations between sulfate and sulfide of 17.3 ± 0.5‰ or 12.6 ± 0.5‰ were recorded for the wildtype or mutant, respectively. The enzymatic machinery that produced these different fractionations was revealed by quantitative proteomics. Results are consistent with a cellular-level response that throttled the supply of electrons and sulfur supply through the sulfate reduction pathway more in the mutant relative to the wildtype, independent of rate. We conclude that the smaller fractionation observed in the mutant strain is a consequence of sulfate reduction that proceeded at a rate that consumed a greater proportion of the strains overall capacity for sulfate reduction. These observations have consequences for models of sulfate reducer metabolism and how it yields different isotopic fractionations, notably, the role of DsrC in central energy metabolism.
Keywords: chemostat; microbial energy metabolism; microbial sulfate reduction; proteomics; sulfur isotope fractionation.
Figures



Similar articles
-
Fractionation of sulfur and hydrogen isotopes in Desulfovibrio vulgaris with perturbed DsrC expression.FEMS Microbiol Lett. 2016 Oct;363(20):fnw226. doi: 10.1093/femsle/fnw226. Epub 2016 Oct 3. FEMS Microbiol Lett. 2016. PMID: 27702753
-
Energy flux couples sulfur isotope fractionation to proteomic and metabolite profiles in Desulfovibrio vulgaris.Geobiology. 2024 May-Jun;22(3):e12600. doi: 10.1111/gbi.12600. Geobiology. 2024. PMID: 38725144
-
DsrC is involved in fermentative growth and interacts directly with the FlxABCD-HdrABC complex in Desulfovibrio vulgaris Hildenborough.Environ Microbiol. 2023 May;25(5):962-976. doi: 10.1111/1462-2920.16335. Epub 2023 Jan 17. Environ Microbiol. 2023. PMID: 36602077
-
What Controls the Sulfur Isotope Fractionation during Dissimilatory Sulfate Reduction?ACS Environ Au. 2023 Jan 3;3(2):76-86. doi: 10.1021/acsenvironau.2c00059. eCollection 2023 Mar 15. ACS Environ Au. 2023. PMID: 37102088 Free PMC article. Review.
-
The "bacterial heterodisulfide" DsrC is a key protein in dissimilatory sulfur metabolism.Biochim Biophys Acta. 2014 Jul;1837(7):1148-64. doi: 10.1016/j.bbabio.2014.03.007. Epub 2014 Mar 22. Biochim Biophys Acta. 2014. PMID: 24662917 Review.
Cited by
-
Influence of Copper on Oleidesulfovibrio alaskensis G20 Biofilm Formation.Microorganisms. 2024 Aug 23;12(9):1747. doi: 10.3390/microorganisms12091747. Microorganisms. 2024. PMID: 39338422 Free PMC article.
-
Integration of text mining and biological network analysis: Identification of essential genes in sulfate-reducing bacteria.Front Microbiol. 2023 Apr 13;14:1086021. doi: 10.3389/fmicb.2023.1086021. eCollection 2023. Front Microbiol. 2023. PMID: 37125195 Free PMC article.
-
Text-Mining to Identify Gene Sets Involved in Biocorrosion by Sulfate-Reducing Bacteria: A Semi-Automated Workflow.Microorganisms. 2023 Jan 3;11(1):119. doi: 10.3390/microorganisms11010119. Microorganisms. 2023. PMID: 36677411 Free PMC article.
References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
-
- Bontognali T. R., Sessions A. L., Allwood A. C., Fischer W. W., Grotzinger J. P., Summons R. E., et al. (2012). Sulfur isotopes of organic matter preserved in 3.45-billion-year-old stromatolites reveal microbial metabolism. Proc. Natl. Acad. Sci. U.S.A. 109 15146–15151. 10.1073/pnas.1207491109 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

