New Insight Into Avian Papillomavirus Ecology and Evolution From Characterization of Novel Wild Bird Papillomaviruses
- PMID: 31031718
- PMCID: PMC6473165
- DOI: 10.3389/fmicb.2019.00701
New Insight Into Avian Papillomavirus Ecology and Evolution From Characterization of Novel Wild Bird Papillomaviruses
Abstract
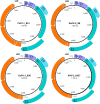
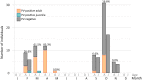
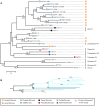
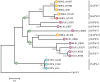
Viruses in the family Papillomaviridae have circular dsDNA genomes of approximately 5.7-8.6 kb that are packaged within non-enveloped, icosahedral capsids. The known papillomavirus (PV) representatives infect vertebrates, and there are currently more than 130 recognized PV species in more than 50 genera. We identified 12 novel avian papillomavirus (APV) types in wild birds that could represent five distinct species and two genera. Viruses were detected in paired oropharyngeal/cloacal swabs collected from six bird species, increasing the number of avian species known to harbor PVs by 40%. A new duck PV (DuPV-3) was found in mallard and American black duck (27.6% estimated prevalence) that was monophyletic with other known DuPVs. A single viral type was identified in Atlantic puffin (PuPV-1, 9.8% estimated prevalence), while a higher genetic diversity was found in other Charadriiformes. Specifically, three types [gull PV-1 (GuPV-1), -2, and -3] were identified in two gull species (estimated prevalence of 17% and 2.6% in American herring and great black-backed gull, respectively), and seven types [kittiwake PV-1 (KiPV-1) through -7] were found in black-legged kittiwake (81.3% estimated prevalence). Significantly higher DuPV-3 circulation was observed in spring compared to fall and in adults compared to juveniles. The studied host species' tendencies to be in crowded environments likely affect infection rates and their migratory behaviors could explain the high viral diversity, illustrating how host behavior can influence viral ecology and distribution. For DuPV-3, GuPV-1, PuPV-1, and KiPV-2, we obtained the complete genomic sequences, which showed the same organization as other known APVs. Phylogenetic analyses showed evidence for virus-host co-divergence at the host taxonomic levels of family, order, and inter-order, but we also observed that host-specificity constraints are relaxed among highly related hosts as we found cross-species transmission within ducks and within gulls. Furthermore, the phylogeny of viruses infecting the Charadriiformes did not match the host phylogeny and gull viruses formed distinct monophyletic clades with kittiwake viruses, possibly reflecting past host-switching events. Considering the vast PV genotype diversity in other hosts and the large number of bird species, many more APVs likely remain to be discovered.
Keywords: avian papillomavirus; molecular epidemiology; papillomavirus; viral ecology; virus discovery; virus evolution.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

