Distinct and Specific Role of NlpC/P60 Endopeptidases LytA and LytB in Cell Elongation and Division of Lactobacillus plantarum
- PMID: 31031721
- PMCID: PMC6473061
- DOI: 10.3389/fmicb.2019.00713
Distinct and Specific Role of NlpC/P60 Endopeptidases LytA and LytB in Cell Elongation and Division of Lactobacillus plantarum
Abstract
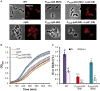
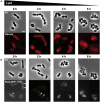
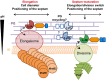
Peptidoglycan (PG) is an essential lattice of the bacterial cell wall that needs to be continuously remodeled to allow growth. This task is ensured by the concerted action of PG synthases that insert new material in the pre-existing structure and PG hydrolases (PGHs) that cleave the PG meshwork at critical sites for its processing. Contrasting with Bacillus subtilis that contains more than 35 PGHs, Lactobacillus plantarum is a non-sporulating rod-shaped bacterium that is predicted to possess a minimal set of 12 PGHs. Their role in morphogenesis and cell cycle remains mostly unexplored, except for the involvement of the glucosaminidase Acm2 in cell separation and the NlpC/P60 D, L-endopeptidase LytA in cell shape maintenance. Besides LytA, L. plantarum encodes three additional NlpC/P60 endopeptidases (i.e., LytB, LytC and LytD). The in silico analysis of these four endopeptidases suggests that they could have redundant functions based on their modular organization, forming two pairs of paralogous enzymes. In this work, we investigate the role of each Lyt endopeptidase in cell morphogenesis in order to evaluate their distinct or redundant functions, and eventually their synthetic lethality. We show that the paralogous LytC and LytD enzymes are not required for cell shape maintenance, which may indicate an accessory role such as in PG recycling. In contrast, LytA and LytB appear to be key players of the cell cycle. We show here that LytA is required for cell elongation while LytB is involved in the spatio-temporal regulation of cell division. In addition, both PGHs are involved in the proper positioning of the division site. The absence of LytA activity is responsible for the asymmetrical positioning of septa in round cells while the lack of LytB results in a lateral misplacement of division planes in rod-shaped cells. Finally, we show that the co-inactivation of LytA and LytB is synthetically affecting cell growth, which confirms the key roles played by both enzymes in PG remodeling during the cell cycle of L. plantarum. Based on the large distribution of NlpC/P60 endopeptidases in low-GC Gram-positive bacteria, these enzymes are attractive targets for the discovery of novel antimicrobial compounds.
Keywords: Lactobacillus; NlpC/P60 endopeptidase; cell cycle; cell wall; morphogenesis; muropeptidase; peptidoglycan hydrolase.
Figures





Similar articles
-
Peptidoglycan NlpC/P60 peptidases in bacterial physiology and host interactions.Cell Chem Biol. 2023 May 18;30(5):436-456. doi: 10.1016/j.chembiol.2022.11.001. Epub 2022 Nov 22. Cell Chem Biol. 2023. PMID: 36417916 Free PMC article. Review.
-
Identification of key peptidoglycan hydrolases for morphogenesis, autolysis, and peptidoglycan composition of Lactobacillus plantarum WCFS1.Microb Cell Fact. 2012 Oct 15;11:137. doi: 10.1186/1475-2859-11-137. Microb Cell Fact. 2012. PMID: 23066986 Free PMC article.
-
Insights into Substrate Specificity of NlpC/P60 Cell Wall Hydrolases Containing Bacterial SH3 Domains.mBio. 2015 Sep 15;6(5):e02327-14. doi: 10.1128/mBio.02327-14. mBio. 2015. PMID: 26374125 Free PMC article.
-
A Secreted NlpC/P60 Endopeptidase from Photobacterium damselae subsp. piscicida Cleaves the Peptidoglycan of Potentially Competing Bacteria.mSphere. 2021 Feb 3;6(1):e00736-20. doi: 10.1128/mSphere.00736-20. mSphere. 2021. PMID: 33536321 Free PMC article.
-
The Cell Wall Hydrolytic NlpC/P60 Endopeptidases in Mycobacterial Cytokinesis: A Structural Perspective.Cells. 2019 Jun 18;8(6):609. doi: 10.3390/cells8060609. Cells. 2019. PMID: 31216697 Free PMC article. Review.
Cited by
-
Cell wall homeostasis in lactic acid bacteria: threats and defences.FEMS Microbiol Rev. 2020 Sep 1;44(5):538-564. doi: 10.1093/femsre/fuaa021. FEMS Microbiol Rev. 2020. PMID: 32495833 Free PMC article. Review.
-
Peptidoglycan NlpC/P60 peptidases in bacterial physiology and host interactions.Cell Chem Biol. 2023 May 18;30(5):436-456. doi: 10.1016/j.chembiol.2022.11.001. Epub 2022 Nov 22. Cell Chem Biol. 2023. PMID: 36417916 Free PMC article. Review.
-
Distinct Amino Acid Availability-Dependent Regulatory Mechanisms of MepS and MepM Levels in Escherichia coli.Front Microbiol. 2021 Jun 30;12:677739. doi: 10.3389/fmicb.2021.677739. eCollection 2021. Front Microbiol. 2021. PMID: 34276609 Free PMC article.
-
Characterization and genomic analysis of phage vB_ValR_NF, representing a new viral family prevalent in the Ulva prolifera blooms.Front Microbiol. 2023 May 5;14:1161265. doi: 10.3389/fmicb.2023.1161265. eCollection 2023. Front Microbiol. 2023. PMID: 37213492 Free PMC article.
-
Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health.Microb Cell Fact. 2020 Nov 7;19(1):203. doi: 10.1186/s12934-020-01464-4. Microb Cell Fact. 2020. PMID: 33160356 Free PMC article. Review.
References
-
- Barrett J. F., Dolinger D. L., Schramm V. L., Shockman G. D. (1984). The mechanism of soluble peptidoglycan hydrolysis by an autolytic muramidase. A processive exodisaccharidase. J. Biol. Chem. 259, 11818–11827. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

