Environmental and Clinical Strains of Vibrio cholerae Non-O1, Non-O139 From Germany Possess Similar Virulence Gene Profiles
- PMID: 31031724
- PMCID: PMC6474259
- DOI: 10.3389/fmicb.2019.00733
Environmental and Clinical Strains of Vibrio cholerae Non-O1, Non-O139 From Germany Possess Similar Virulence Gene Profiles
Abstract
Vibrio cholerae is a natural inhabitant of aquatic ecosystems globally. Strains of the serogroups O1 and O139 cause the epidemic diarrheal disease cholera. In Northern European waters, V. cholerae bacteria belonging to other serogroups (designated non-O1, non-O139) are present, of which some strains have been associated with gastrointestinal infections or extraintestinal infections, like wound infections or otitis. For this study, environmental strains from the German coastal waters of the North Sea and the Baltic Sea were selected (100 strains) and compared to clinical strains (10 isolates) that were from patients who contracted the infections in the same geographical region. The strains were characterized by MLST and examined by PCR for the presence of virulence genes encoding the cholera toxin, the toxin-coregulated pilus (TCP), and other virulence-associated accessory factors. The latter group comprised hemolysins, RTX toxins, cholix toxin, pandemic islands, and type III secretion system (TTSS). Phenotypic assays for hemolytic activity against human and sheep erythrocytes were also performed. The results of the MLST analysis revealed a considerable heterogeneity of sequence types (in total 74 STs). The presence of virulence genes was also variable and 30 profiles were obtained by PCR. One profile was found in 38 environmental strains and six clinical strains. Whole genome sequencing (WGS) was performed on 15 environmental and 7 clinical strains that were ST locus variants in one, two, or three alleles. Comparison of WGS results revealed that a set of virulence genes found in some clinical strains is also present in most environmental strains irrespective of the ST. In few strains, more virulence factors are acquired through horizontal gene transfer (i.e., TTSS, genomic islands). A distinction between clinical and environmental strains based on virulence gene profiles is not possible for our strains. Probably, many virulence traits of V. cholerae evolved in response to biotic and abiotic pressure and serve adaptation purposes in the natural aquatic environment, but provide a prerequisite for infection of susceptible human hosts. These findings indicate the need for surveillance of Vibrio spp. in Germany, as due to global warming abundance of Vibrio will rise and infections are predicted to increase.
Keywords: Baltic Sea; Germany; North Sea; Vibrio cholerae; clinical isolates; multilocus sequence typing; virulence-associated factors; whole genome sequencing.
Figures
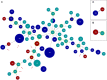


 ). SNP tree was conducted using CSI Phylogeny 1.4 under default settings and the exclusion of heterozygous SNPs. Single nucleotide polymorphisms (SNPs) were called by mapping to the V. cholerae VN-00533 genome as reference (MWZM00000000). Scale bar represents the number of nucleotide substitutions per site and numbers indicate branch length. Basic information about the sample type and the year of isolation are given after the strain identifier: North Sea/seawater (NS-sw), North Sea/bivalve mollusk (NS-bm), Baltic Sea/seawater (BS-sw), and peritoneal fluid (pf).
). SNP tree was conducted using CSI Phylogeny 1.4 under default settings and the exclusion of heterozygous SNPs. Single nucleotide polymorphisms (SNPs) were called by mapping to the V. cholerae VN-00533 genome as reference (MWZM00000000). Scale bar represents the number of nucleotide substitutions per site and numbers indicate branch length. Basic information about the sample type and the year of isolation are given after the strain identifier: North Sea/seawater (NS-sw), North Sea/bivalve mollusk (NS-bm), Baltic Sea/seawater (BS-sw), and peritoneal fluid (pf).Similar articles
-
Genetic and phenotypic analysis of Vibrio cholerae non-O1, non-O139 isolated from German and Austrian patients.Eur J Clin Microbiol Infect Dis. 2014 May;33(5):767-78. doi: 10.1007/s10096-013-2011-9. Epub 2013 Nov 10. Eur J Clin Microbiol Infect Dis. 2014. PMID: 24213848 Free PMC article.
-
Characterization and Genetic Variation of Vibrio cholerae Isolated from Clinical and Environmental Sources in Thailand.PLoS One. 2017 Jan 19;12(1):e0169324. doi: 10.1371/journal.pone.0169324. eCollection 2017. PLoS One. 2017. PMID: 28103259 Free PMC article.
-
Phenotypic and Genotypic Properties of Vibrio cholerae non-O1, non-O139 Isolates Recovered from Domestic Ducks in Germany.Microorganisms. 2020 Jul 23;8(8):1104. doi: 10.3390/microorganisms8081104. Microorganisms. 2020. PMID: 32717968 Free PMC article.
-
Epidemiology & molecular biology of Vibrio cholerae O139 Bengal.Indian J Med Res. 1996 Jul;104:14-27. Indian J Med Res. 1996. PMID: 8783504 Review.
-
Emergence of a new cholera pandemic: molecular analysis of virulence determinants in Vibrio cholerae O139 and development of a live vaccine prototype.J Infect Dis. 1994 Aug;170(2):278-83. doi: 10.1093/infdis/170.2.278. J Infect Dis. 1994. PMID: 8035010 Review.
Cited by
-
Virulence and resistance patterns of Vibrio cholerae non-O1/non-O139 acquired in Germany and other European countries.Front Microbiol. 2023 Nov 22;14:1282135. doi: 10.3389/fmicb.2023.1282135. eCollection 2023. Front Microbiol. 2023. PMID: 38075873 Free PMC article.
-
Vibrio cholerae Infection Induces Strain-Specific Modulation of the Zebrafish Intestinal Microbiome.Infect Immun. 2021 Aug 16;89(9):e0015721. doi: 10.1128/IAI.00157-21. Epub 2021 Aug 16. Infect Immun. 2021. PMID: 34061623 Free PMC article.
-
Incidence and Virulence Factor Profiling of Vibrio Species: A Study on Hospital and Community Wastewater Effluents.Microorganisms. 2023 Sep 29;11(10):2449. doi: 10.3390/microorganisms11102449. Microorganisms. 2023. PMID: 37894107 Free PMC article.
-
Cholera Dynamics and the Emergence of Pandemic Vibrio cholerae.Adv Exp Med Biol. 2023;1404:127-147. doi: 10.1007/978-3-031-22997-8_7. Adv Exp Med Biol. 2023. PMID: 36792874
-
Minority report: small-scale metagenomic analysis of the non-bacterial kitchen sponge microbiota.Arch Microbiol. 2022 Jun 4;204(7):363. doi: 10.1007/s00203-022-02969-9. Arch Microbiol. 2022. PMID: 35661258 Free PMC article.
References
-
- Alm R. A., Stroeher U. H., Manning P. A. (1988). Extracellular proteins of Vibrio cholerae: nucleotide sequence of the structural gene (hlyA) for the haemolysin of the haemolytic El Tor strain 017 and characterization of the hlyA mutation in the non-haemolytic classical strain 569B. Mol. Microbiol. 2 481–488. 10.1111/j.1365-2958.1988.tb00054.x - DOI - PubMed
-
- Baker-Austin C., Trinanes J. A., Taylor N. G. H., Hartnell R., Siitonen A., Martinez-Urtaza J. (2012). Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Chang. 3 73–77. 10.1038/nclimate1628 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

