Epigenetic DNA Modifications Are Correlated With B Chromosomes and Sex in the Cichlid Astatotilapia latifasciata
- PMID: 31031803
- PMCID: PMC6474290
- DOI: 10.3389/fgene.2019.00324
Epigenetic DNA Modifications Are Correlated With B Chromosomes and Sex in the Cichlid Astatotilapia latifasciata
Abstract
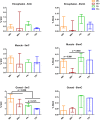
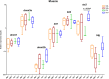
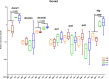
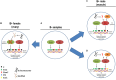
Supernumerary B chromosomes are dispensable elements found in several groups of eukaryotes, and their impacts in host organisms are not clear. The cichlid fish Astatotilapia latifasciata presents one or two large metacentric B chromosomes. These elements affect the transcription of several classes of RNAs. Here, we evaluated the epigenetic DNA modification status of B chromosomes using immunocytogenetics and assessed the impact of B chromosome presence on the global contents of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) and the molecular mechanisms underlying these variations. We found that the B chromosome of A. latifasciata has an active pattern of DNA epimarks, and its presence promotes the loss of 5mC in gonads of females with B chromosome (FB+) and promotes the loss of 5hmC in the muscle of males with the B element (MB+). Based on the transcriptional quantification of DNA modification genes (dnmt, tet, and tdg) and their candidate regulators (idh genes, microRNAs, and long non-coding RNAs) and on RNA-protein interaction prediction, we suggest the occurrence of passive demethylation in gonads of FB+ and 5hmC loss by Tet inhibition or by 5hmC oxidation in MB+ muscle. We suggest that these results can also explain the previously reported variations in the transcription levels of several classes of RNA depending on B chromosome presence. The DNA modifications detected here are also influenced by sex. Although the correlation between B chromosomes and sex has been previously reported, it remains unexplained. The B chromosome of A. latifasciata seems to be active and impacts cell physiology in a very complex way, including at the epigenetic level.
Keywords: 5-hydroxymethylcytosine; 5-methylcytosine; DNA demethylation; DNA methylation; microRNA; supernumerary chromosome.
Figures




Similar articles
-
Meiotic behavior, transmission and active genes of B chromosomes in the cichlid Astatotilapia latifasciata: new clues about nature, evolution and maintenance of accessory elements.Mol Genet Genomics. 2022 Jul;297(4):1151-1167. doi: 10.1007/s00438-022-01911-4. Epub 2022 Jun 15. Mol Genet Genomics. 2022. PMID: 35704117
-
Landscape of Transposable Elements Focusing on the B Chromosome of the Cichlid Fish Astatotilapia latifasciata.Genes (Basel). 2018 May 23;9(6):269. doi: 10.3390/genes9060269. Genes (Basel). 2018. PMID: 29882892 Free PMC article.
-
The repetitive DNA element BncDNA, enriched in the B chromosome of the cichlid fish Astatotilapia latifasciata, transcribes a potentially noncoding RNA.Chromosoma. 2017 Mar;126(2):313-323. doi: 10.1007/s00412-016-0601-x. Epub 2016 May 12. Chromosoma. 2017. PMID: 27169573
-
Epigenetic regulation in human melanoma: past and future.Epigenetics. 2015;10(2):103-21. doi: 10.1080/15592294.2014.1003746. Epigenetics. 2015. PMID: 25587943 Free PMC article. Review.
-
TET Methylcytosine Oxidases in T Cell and B Cell Development and Function.Front Immunol. 2017 Mar 31;8:220. doi: 10.3389/fimmu.2017.00220. eCollection 2017. Front Immunol. 2017. PMID: 28408905 Free PMC article. Review.
Cited by
-
Meiotic behavior, transmission and active genes of B chromosomes in the cichlid Astatotilapia latifasciata: new clues about nature, evolution and maintenance of accessory elements.Mol Genet Genomics. 2022 Jul;297(4):1151-1167. doi: 10.1007/s00438-022-01911-4. Epub 2022 Jun 15. Mol Genet Genomics. 2022. PMID: 35704117
-
A genomic glimpse of B chromosomes in cichlids.Genes Genomics. 2021 Mar;43(3):199-208. doi: 10.1007/s13258-021-01049-4. Epub 2021 Feb 6. Genes Genomics. 2021. PMID: 33547625 Review.
-
Differential expression of miRNAs in the presence of B chromosome in the cichlid fish Astatotilapia latifasciata.BMC Genomics. 2021 May 12;22(1):344. doi: 10.1186/s12864-021-07651-w. BMC Genomics. 2021. PMID: 33980143 Free PMC article.
-
First characterization of PIWI-interacting RNA clusters in a cichlid fish with a B chromosome.BMC Biol. 2022 Sep 21;20(1):204. doi: 10.1186/s12915-022-01403-2. BMC Biol. 2022. PMID: 36127679 Free PMC article.
References
-
- Arvinden V. R., Rao A. K. D. M., Rajkumar T., Mani S. (2017). Regulation and functional significance of 5-hydroxymethylcytosine in cancer. Epigenomes 1:19 10.3390/epigenomes1030019 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

