An aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome
- PMID: 31032404
- PMCID: PMC6482013
- DOI: 10.1126/sciadv.aav1110
An aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome
Abstract
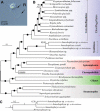
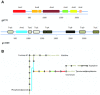
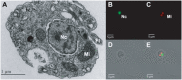
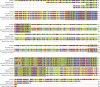
Dinoflagellates are microbial eukaryotes that have exceptionally large nuclear genomes; however, their organelle genomes are small and fragmented and contain fewer genes than those of other eukaryotes. The genus Amoebophrya (Syndiniales) comprises endoparasites with high genetic diversity that can infect other dinoflagellates, such as those forming harmful algal blooms (e.g., Alexandrium). We sequenced the genome (~100 Mb) of Amoebophrya ceratii to investigate the early evolution of genomic characters in dinoflagellates. The A. ceratii genome encodes almost all essential biosynthetic pathways for self-sustaining cellular metabolism, suggesting a limited dependency on its host. Although dinoflagellates are thought to have descended from a photosynthetic ancestor, A. ceratii appears to have completely lost its plastid and nearly all genes of plastid origin. Functional mitochondria persist in all life stages of A. ceratii, but we found no evidence for the presence of a mitochondrial genome. Instead, all mitochondrial proteins appear to be lost or encoded in the A. ceratii nucleus.
Figures

References
-
- Keeling P. J., Burger G., Durnford D. G., Lang B. F., Lee R. W., Pearlman R. E., Roger A. J., Gray M. W., The tree of eukaryotes. Trends Ecol. Evol. 20, 670–676 (2005). - PubMed
-
- Lin S., Genomic understanding of dinoflagellates. Res. Microbiol. 162, 551–569 (2011). - PubMed
-
- Gornik S. G., Ford K. L., Mulhern T. D., Bacic A., McFadden G. I., Waller R. F., Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr. Biol. 22, 2303–2312 (2012). - PubMed
-
- LaJeunesse T. C., Lambert G., Andersen R. A., Coffroth M. A., Galbraith D. W., Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. J. Phycol. 41, 880–886 (2005).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

