The MACADAM database: a MetAboliC pAthways DAtabase for Microbial taxonomic groups for mining potential metabolic capacities of archaeal and bacterial taxonomic groups
- PMID: 31032842
- PMCID: PMC6487390
- DOI: 10.1093/database/baz049
The MACADAM database: a MetAboliC pAthways DAtabase for Microbial taxonomic groups for mining potential metabolic capacities of archaeal and bacterial taxonomic groups
Abstract
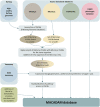
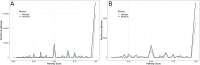
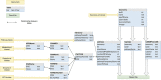
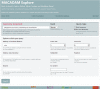
Progress in genome sequencing and bioinformatics opens up new possibilities, including that of correlating genome annotations with functional information such as metabolic pathways. Thanks to the development of functional annotation databases, scientists are able to link genome annotations with functional annotations. We present MetAboliC pAthways DAtabase for Microbial taxonomic groups (MACADAM) here, a user-friendly database that makes it possible to find presence/absence/completeness statistics for metabolic pathways at a given microbial taxonomic position. For each prokaryotic 'RefSeq complete genome', MACADAM builds a pathway genome database (PGDB) using Pathway Tools software based on MetaCyc data that includes metabolic pathways as well as associated metabolites, reactions and enzymes. To ensure the highest quality of the genome functional annotation data, MACADAM also contains MicroCyc, a manually curated collection of PGDBs; Functional Annotation of Prokaryotic Taxa (FAPROTAX), a manually curated functional annotation database; and the IJSEM phenotypic database. The MACADAM database contains 13 509 PGDBs (13 195 bacterial and 314 archaeal), 1260 unique metabolic pathways, completed with 82 functional annotations from FAPROTAX and 16 from the IJSEM phenotypic database. MACADAM contains a total of 7921 metabolites, 592 enzymatic reactions, 2134 EC numbers and 7440 enzymes. MACADAM can be queried at any rank of the NCBI taxonomy (from phyla to species). It provides the possibility to explore functional information completed with metabolites, enzymes, enzymatic reactions and EC numbers. MACADAM returns a tabulated file containing a list of pathways with two scores (pathway score and pathway frequency score) that are present in the queried taxa. The file also contains the names of the organisms in which the pathways are found and the metabolic hierarchy associated with the pathways. Finally, MACADAM can be downloaded as a single file and queried with SQLite or python command lines or explored through a web interface.
© The Author(s) 2019. Published by Oxford University Press.
Figures



References
-
- Bergey D.H., Buchanan R.E., Gibbons N.E. et al. (1974) Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins Co, Baltimore.
-
- Garrity G., Boone D.R. and Castenholz R.W. (eds) (2001) Bergey’s Manual of Systematic Bacteriology: The Archaea and the Deeply Branching and Phototrophic Bacteria, Vol. 1, 2nd edn. Springer-Verlag, New York.
-
- Garrity G. (ed) (2005) Bergey’s Manual of Systematic Bacteriology: The Proteobacteria, Vol. 2, 2nd edn. Springer, USA.
-
- Vos P., Garrity G., Jones D. et al. (eds) (2009) Bergey’s Manual of Systematic Bacteriology: The Firmicutes, Vol. 3, 2nd edn. Springer-Verlag, New York.
-
- Krieg N.R., Ludwig W., Whitman W. et al. (eds) (2010) Bergey’s Manual of Systematic Bacteriology: The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes, Vol. 4, 2nd edn. Springer-Verlag, New York.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

