Host metabolites stimulate the bacterial proton motive force to enhance the activity of aminoglycoside antibiotics
- PMID: 31034512
- PMCID: PMC6508747
- DOI: 10.1371/journal.ppat.1007697
Host metabolites stimulate the bacterial proton motive force to enhance the activity of aminoglycoside antibiotics
Abstract
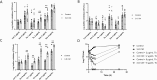
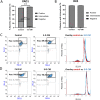
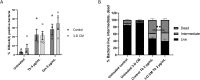
Antibiotic susceptibility of bacterial pathogens is typically evaluated using in vitro assays that do not consider the complex host microenvironment. This may help explaining a significant discrepancy between antibiotic efficacy in vitro and in vivo, with some antibiotics being effective in vitro but not in vivo or vice versa. Nevertheless, it is well-known that antibiotic susceptibility of bacteria is driven by environmental factors. Lung epithelial cells enhance the activity of aminoglycoside antibiotics against the opportunistic pathogen Pseudomonas aeruginosa, yet the mechanism behind is unknown. The present study addresses this gap and provides mechanistic understanding on how lung epithelial cells stimulate aminoglycoside activity. To investigate the influence of the local host microenvironment on antibiotic activity, an in vivo-like three-dimensional (3-D) lung epithelial cell model was used. We report that conditioned medium of 3-D lung cells, containing secreted but not cellular components, potentiated the bactericidal activity of aminoglycosides against P. aeruginosa, including resistant clinical isolates, and several other pathogens. In contrast, conditioned medium obtained from the same cell type, but grown as conventional (2-D) monolayers did not influence antibiotic efficacy. We found that 3-D lung cells secreted endogenous metabolites (including succinate and glutamate) that enhanced aminoglycoside activity, and provide evidence that bacterial pyruvate metabolism is linked to the observed potentiation of antimicrobial activity. Biochemical and phenotypic assays indicated that 3-D cell conditioned medium stimulated the proton motive force (PMF), resulting in increased bacterial intracellular pH. The latter stimulated antibiotic uptake, as determined using fluorescently labelled tobramycin in combination with flow cytometry analysis. Our findings reveal a cross-talk between host and bacterial metabolic pathways, that influence downstream activity of antibiotics. Understanding the underlying basis of the discrepancy between the activity of antibiotics in vitro and in vivo may lead to improved diagnostic approaches and pave the way towards novel means to stimulate antibiotic activity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






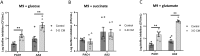
Similar articles
-
Rapid Freezing Enables Aminoglycosides To Eradicate Bacterial Persisters via Enhancing Mechanosensitive Channel MscL-Mediated Antibiotic Uptake.mBio. 2020 Feb 11;11(1):e03239-19. doi: 10.1128/mBio.03239-19. mBio. 2020. PMID: 32047133 Free PMC article.
-
Influence of three-dimensional lung epithelial cells and interspecies interactions on antibiotic efficacy against Mycobacterium abscessus and Pseudomonas aeruginosa.Pathog Dis. 2018 Jun 1;76(4). doi: 10.1093/femspd/fty034. Pathog Dis. 2018. PMID: 29648588
-
Antimicrobial efficacy against Pseudomonas aeruginosa biofilm formation in a three-dimensional lung epithelial model and the influence of fetal bovine serum.Sci Rep. 2017 Mar 3;7:43321. doi: 10.1038/srep43321. Sci Rep. 2017. PMID: 28256611 Free PMC article.
-
Tobramycin: a review of its antibacterial and pharmacokinetic properties and therapeutic use.Drugs. 1976;12(3):166-200. doi: 10.2165/00003495-197612030-00002. Drugs. 1976. PMID: 789045 Review.
-
A mini-review: environmental and metabolic factors affecting aminoglycoside efficacy.World J Microbiol Biotechnol. 2022 Nov 9;39(1):7. doi: 10.1007/s11274-022-03445-8. World J Microbiol Biotechnol. 2022. PMID: 36350431 Free PMC article. Review.
Cited by
-
Potentiation of Aminoglycoside Lethality by C4-Dicarboxylates Requires RpoN in Antibiotic-Tolerant Pseudomonas aeruginosa.Antimicrob Agents Chemother. 2019 Sep 23;63(10):e01313-19. doi: 10.1128/AAC.01313-19. Print 2019 Oct. Antimicrob Agents Chemother. 2019. PMID: 31383655 Free PMC article.
-
Aminoglycoside uptake, stress, and potentiation in Gram-negative bacteria: new therapies with old molecules.Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0003622. doi: 10.1128/mmbr.00036-22. Epub 2023 Dec 4. Microbiol Mol Biol Rev. 2023. PMID: 38047635 Free PMC article. Review.
-
Bioenergetic suppression by redox-active metabolites promotes antibiotic tolerance in Pseudomonas aeruginosa.Proc Natl Acad Sci U S A. 2024 Nov 12;121(46):e2406555121. doi: 10.1073/pnas.2406555121. Epub 2024 Nov 6. Proc Natl Acad Sci U S A. 2024. PMID: 39503891 Free PMC article.
-
Metabolic state-driven nutrient-based approach to combat bacterial antibiotic resistance.NPJ Antimicrob Resist. 2025 Apr 4;3(1):24. doi: 10.1038/s44259-025-00092-5. NPJ Antimicrob Resist. 2025. PMID: 40185857 Free PMC article. Review.
-
Unlocking Enhanced Efficacy of Aminoglycoside Antibiotics Against Pseudomonas aeruginosa.Microb Biotechnol. 2025 Jun;18(6):e70174. doi: 10.1111/1751-7915.70174. Microb Biotechnol. 2025. PMID: 40448301 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

