OsMADS18, a membrane-bound MADS-box transcription factor, modulates plant architecture and the abscisic acid response in rice
- PMID: 31034557
- PMCID: PMC6685668
- DOI: 10.1093/jxb/erz198
OsMADS18, a membrane-bound MADS-box transcription factor, modulates plant architecture and the abscisic acid response in rice
Abstract
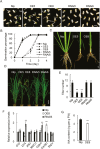
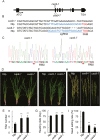
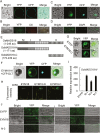
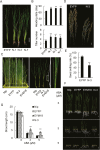
The APETALA1 (AP1)/FRUITFULL (FUL)-like transcription factor OsMADS18 plays diverse functions in rice development, but the underlying molecular mechanisms are far from fully understood. Here, we report that down-regulation of OsMADS18 expression in RNAi lines caused a delay in seed germination and young seedling growth, whereas the overexpression of OsMADS18 produced plants with fewer tillers. In targeted OsMADS18 genome-edited mutants (osmads18-cas9), an increased number of tillers, altered panicle size, and reduced seed setting were observed. The EYFP-OsMADS18 (full-length) protein was localized to the nucleus and plasma membrane but the EYFP-OsMADS18-N (N-terminus) protein mainly localized to the nucleus. The expression of OsMADS18 could be stimulated by abscisic acid (ABA), and ABA stimulation triggered the cleavage of HA-OsMADS18 and the translocation of OsMADS18 from the plasma membrane to the nucleus. The inhibitory effect of ABA on seedling growth was less effective in the OsMADS18-overexpressing plants. The expression of a set of ABA-responsive genes was significantly reduced in the overexpressing plants. The phenotypes of transgenic plants expressing EYFP-OsMADS18-N resembled those observed in the osmads18-cas9 mutants. Analysis of the interaction of OsMADS18 with OsMADS14, OsMADS15, and OsMADS57 strongly suggests an essential role for OsMADS18 in rice development.
Keywords: ABA; MADS-box; membrane-bound transcription factor; plant architecture; rice; tiller.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures


References
-
- Amir Hossain M, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB. 2010. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Molecular Biology 72, 557–566. - PubMed
-
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. 2009. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant & Cell Physiology 50, 1416–1424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

