Protein Hydroxylation by Hypoxia-Inducible Factor (HIF) Hydroxylases: Unique or Ubiquitous?
- PMID: 31035491
- PMCID: PMC6562979
- DOI: 10.3390/cells8050384
Protein Hydroxylation by Hypoxia-Inducible Factor (HIF) Hydroxylases: Unique or Ubiquitous?
Abstract
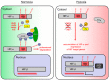
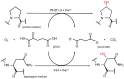
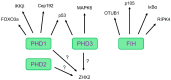
All metazoans that utilize molecular oxygen (O2) for metabolic purposes have the capacity to adapt to hypoxia, the condition that arises when O2 demand exceeds supply. This is mediated through activation of the hypoxia-inducible factor (HIF) pathway. At physiological oxygen levels (normoxia), HIF-prolyl hydroxylases (PHDs) hydroxylate proline residues on HIF-α subunits leading to their destabilization by promoting ubiquitination by the von-Hippel Lindau (VHL) ubiquitin ligase and subsequent proteasomal degradation. HIF-α transactivation is also repressed in an O2-dependent way due to asparaginyl hydroxylation by the factor-inhibiting HIF (FIH). In hypoxia, the O2-dependent hydroxylation of HIF-α subunits by PHDs and FIH is reduced, resulting in HIF-α accumulation, dimerization with HIF-β and migration into the nucleus to induce an adaptive transcriptional response. Although HIFs are the canonical substrates for PHD- and FIH-mediated protein hydroxylation, increasing evidence indicates that these hydroxylases may also have alternative targets. In addition to PHD-conferred alterations in protein stability, there is now evidence that hydroxylation can affect protein activity and protein/protein interactions for alternative substrates. PHDs can be pharmacologically inhibited by a new class of drugs termed prolyl hydroxylase inhibitors which have recently been approved for the treatment of anemia associated with chronic kidney disease. The identification of alternative targets of HIF hydroxylases is important in order to fully elucidate the pharmacology of hydroxylase inhibitors (PHI). Despite significant technical advances, screening, detection and verification of alternative functional targets for PHDs and FIH remain challenging. In this review, we discuss recently proposed non-HIF targets for PHDs and FIH and provide an overview of the techniques used to identify these.
Keywords: Cep192; FOXO3a; HIF-prolyl hydroxylases; IKK-β; MAPK6; OTUB1; RIPK4; factor inhibiting HIF; hypoxia; hypoxia-inducible factor; mass spectrometry; p105; p53; prolyl hydroxylation.
Conflict of interest statement
C.T.T. is a member of the Scientific Advisory Board of Akebia Therapeutics.
Figures
References
-
- Zhang H., Gao P., Fukuda R., Kumar G., Krishnamachary B., Zeller K.I., Dang C.V., Semenza G.L. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

