How Ricin Damages the Ribosome
- PMID: 31035546
- PMCID: PMC6562825
- DOI: 10.3390/toxins11050241
How Ricin Damages the Ribosome
Abstract
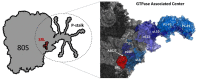
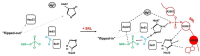
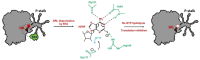
Ricin belongs to the group of ribosome-inactivating proteins (RIPs), i.e., toxins that have evolved to provide particular species with an advantage over other competitors in nature. Ricin possesses RNA N-glycosidase activity enabling the toxin to eliminate a single adenine base from the sarcin-ricin RNA loop (SRL), which is a highly conserved structure present on the large ribosomal subunit in all species from the three domains of life. The SRL belongs to the GTPase associated center (GAC), i.e., a ribosomal element involved in conferring unidirectional trajectory for the translational apparatus at the expense of GTP hydrolysis by translational GTPases (trGTPases). The SRL represents a critical element in the GAC, being the main triggering factor of GTP hydrolysis by trGTPases. Enzymatic removal of a single adenine base at the tip of SRL by ricin blocks GTP hydrolysis and, at the same time, impedes functioning of the translational machinery. Here, we discuss the consequences of SRL depurination by ricin for ribosomal performance, with emphasis on the mechanistic model overview of the SRL modus operandi.
Keywords: GTPase associated center (GAC); P-proteins; ribosome; ribosome-inactivating protein; ricin; sarcin-ricin loop (SRL); translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Barbieri L., Polito L., Bolognesi A., Ciani M., Pelosi E., Farini V., Jha A.K., Sharma N., Vivanco J.M., Chambery A., et al. Ribosome-inactivating proteins in edible plants and purification and characterization of a new ribosome-inactivating protein from Cucurbita moschata. Biochim. Biophys. Acta. 2006;1760:783–792. doi: 10.1016/j.bbagen.2006.01.002. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

