A Preliminary Metagenome Analysis Based on a Combination of Protein Domains
- PMID: 31035705
- PMCID: PMC6630717
- DOI: 10.3390/proteomes7020019
A Preliminary Metagenome Analysis Based on a Combination of Protein Domains
Abstract
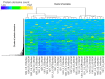
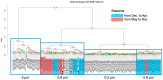
Metagenomic data have mainly been addressed by showing the composition of organisms based on a small part of a well-examined genomic sequence, such as ribosomal RNA genes and mitochondrial DNAs. On the contrary, whole metagenomic data obtained by the shotgun sequence method have not often been fully analyzed through a homology search because the genomic data in databases for living organisms on earth are insufficient. In order to complement the results obtained through homology-search-based methods with shotgun metagenomes data, we focused on the composition of protein domains deduced from the sequences of genomes and metagenomes, and we utilized them in characterizing genomes and metagenomes, respectively. First, we compared the relationships based on similarities in the protein domain composition with the relationships based on sequence similarities. We searched for protein domains of 325 bacterial species produced using the Pfam database. Next, the correlation coefficients of protein domain compositions between every pair of bacteria were examined. Every pairwise genetic distance was also calculated from 16S rRNA or DNA gyrase subunit B. We compared the results of these methods and found a moderate correlation between them. Essentially, the same results were obtained when we used partial random 100 bp DNA sequences of the bacterial genomes, which simulated raw sequence data obtained from short-read next-generation sequences. Then, we applied the method for analyzing the actual environmental data obtained by shotgun sequencing. We found that the transition of the microbial phase occurred because the seasonal change in water temperature was shown by the method. These results showed the usability of the method in characterizing metagenomic data based on protein domain compositions.
Keywords: correlation coefficient; environmental DNA; metagenomics; phylogenetic analysis; protein domain.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

