1H-NMR Metabolite Fingerprinting Analysis Reveals a Disease Biomarker and a Field Treatment Response in Xylella fastidiosa subsp. pauca-Infected Olive Trees
- PMID: 31035723
- PMCID: PMC6571561
- DOI: 10.3390/plants8050115
1H-NMR Metabolite Fingerprinting Analysis Reveals a Disease Biomarker and a Field Treatment Response in Xylella fastidiosa subsp. pauca-Infected Olive Trees
Abstract
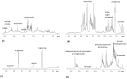
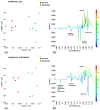
Xylella fastidiosa subsp. pauca is a xylem-limited bacterial phytopathogen currently found associated on many hectares with the "olive quick decline syndrome" in the Apulia region (Southern Italy), and the cultivars Ogliarola salentina and Cellina di Nardò result in being particularly sensitive to the disease. In order to find compounds showing the capability of reducing the population cell density of the pathogen within the leaves, we tested, in some olive orchards naturally-infected by the bacterium, a zinc-copper-citric acid biocomplex, namely Dentamet®, by spraying it to the crown, once per month, during spring and summer. The occurrence of the pathogen in the four olive orchards chosen for the trial was molecularly assessed. A 1H NMR metabolomic approach, in conjunction with a multivariate statistical analysis, was applied to investigate the metabolic pattern of both infected and treated adult olive cultivars, Ogliarola salentina and Cellina di Nardò trees, in two sampling periods, performed during the first year of the trial. For both cultivars and sampling periods, the orthogonal partial least squares discriminant analysis (OPLS-DA) gave good models of separation according to the treatment application. In both cultivars, some metabolites such as quinic acid, the aldehydic form of oleoeuropein, ligstroside and phenolic compounds, were consistently found as discriminative for the untreated olive trees in comparison with the Dentamet®-treated trees. Quinic acid, a precursor of lignin, was confirmed as a disease biomarker for the olive trees infected by X. fastidiosa subsp. pauca. When treated with Dentamet®, the two cultivars showed a distinct response. A consistent increase in malic acid was observed for the Ogliarola salentina trees, whereas in the Cellina di Nardò trees the treatments attenuate the metabolic response to the infection. To note that in Cellina di Nardò trees at the first sampling, an increase in γ-aminobutyric acid (GABA) was observed. This study highlights how the infection incited by X. fastidiosa subsp. pauca strongly modifies the overall metabolism of olive trees, and how a zinc-copper-citric acid biocomplex can induce an early re-programming of the metabolic pathways in the infected trees.
Keywords: GABA; malic acid; metabolomics; multivariate statistical analysis; polyphenols; quinic acid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Schaad N.W., Postnikova E., Lacy G., Fatmi M.B., Chang C.-J. Xylella fastidiosa subspecies: X. fastidiosa subsp piercei, subsp. nov., X. fastidiosa subsp. multiplex subsp. nov., and X. fastidiosa subsp. pauca subsp. nov. Syst. Appl. Microbiol. 2004;27:290–300. - PubMed
-
- Martelli G., Boscia D., Porcelli F., Saponari M. The olive quick decline syndrome in south-east Italy: A threatening phytosanitary emergency. Eur. J. Plant Pathol. 2016;144:235–243. doi: 10.1007/s10658-015-0784-7. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

