Serine-rich repeat proteins from gut microbes
- PMID: 31035824
- PMCID: PMC6973325
- DOI: 10.1080/19490976.2019.1602428
Serine-rich repeat proteins from gut microbes
Abstract
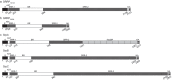
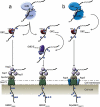
Serine-rich repeat proteins (SRRPs) have emerged as an important group of cell surface adhesins found in a growing number of Gram-positive bacteria. Studies focused on SRRPs from streptococci and staphylococci demonstrated that these proteins are O-glycosylated on serine or threonine residues and exported via an accessory secretion (aSec) system. In pathogens, these adhesins contribute to disease pathogenesis and represent therapeutic targets. Recently, the non-canonical aSec system has been identified in the genomes of gut microbes and characterization of their associated SRRPs is beginning to unfold, showing their role in mediating attachment and biofilm formation. Here we provide an update of the occurrence, structure, and function of SRRPs across bacteria, with emphasis on the molecular and biochemical properties of SRRPs from gut symbionts, particularly Lactobacilli. These emerging studies underscore the range of ligands recognized by these adhesins and the importance of SRRP glycosylation in the interaction of gut microbes with the host.
Keywords: Lactobacillus; Serine rich repeat protein; adhesin; gut symbiont; protein glycosylation.
Figures


References
-
- Wu H, Mintz KP, Ladha M, Fives-Taylor PM. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol. 1998;28:487–500. - PubMed
-
- Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol. 2002;44:1081–1094. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
