Effect of transcription inhibition and generation of suppressive viral non-coding RNAs
- PMID: 31036006
- PMCID: PMC6489247
- DOI: 10.1186/s12977-019-0475-0
Effect of transcription inhibition and generation of suppressive viral non-coding RNAs
Abstract
Background: HIV-1 patients receiving combination antiretroviral therapy (cART) survive infection but require life-long adherence at high expense. In chronic cART-treated patients with undetectable viral titers, cell-associated viral RNA is still detectable, pointing to low-level viral transcriptional leakiness. To date, there are no FDA-approved drugs against HIV-1 transcription. We have previously shown that F07#13, a third generation Tat peptide mimetic with competitive activity against Cdk9/T1-Tat binding sites, inhibits HIV-1 transcription in vitro and in vivo.
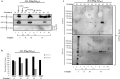
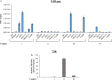
Results: Here, we demonstrate that increasing concentrations of F07#13 (0.01, 0.1, 1 µM) cause a decrease in Tat levels in a dose-dependent manner by inhibiting the Cdk9/T1-Tat complex formation and subsequent ubiquitin-mediated Tat sequestration and degradation. Our data indicate that complexes I and IV contain distinct patterns of ubiquitinated Tat and that transcriptional inhibition induced by F07#13 causes an overall reduction in Tat levels. This reduction may be triggered by F07#13 but ultimately is mediated by TAR-gag viral RNAs that bind suppressive transcription factors (similar to 7SK, NRON, HOTAIR, and Xist lncRNAs) to enhance transcriptional gene silencing and latency. These RNAs complex with PRC2, Sin3A, and Cul4B, resulting in epigenetic modifications. Finally, we observed an F07#13-mediated decrease of viral burden by targeting the R region of the long terminal repeat (HIV-1 promoter region, LTR), promoting both paused polymerases and increased efficiency of CRISPR/Cas9 editing in infected cells. This implies that gene editing may be best performed under a repressed transcriptional state.
Conclusions: Collectively, our results indicate that F07#13, which can terminate RNA Polymerase II at distinct sites, can generate scaffold RNAs, which may assemble into specific sets of "RNA Machines" that contribute to gene regulation. It remains to be seen whether these effects can also be seen in various clades that have varying promoter strength, mutant LTRs, and in patient samples.
Keywords: CRISPR; Gene silencing; HIV-1; Latency; Transcription; cART.
Conflict of interest statement
Authors declare no potential conflicts of interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Figures




References
-
- Akpamagbo YA, DeMarino C, Pleet ML, Schwab A, Rodriguez M, Barclay RA, Sampey G, Iordanskiy S, El-Hage N, Kashanchi F. HIV-1 transcription inhibitors increase the synthesis of viral non-coding RNA that contribute to latency. Curr Pharm Des. 2017;23:4133–4144. doi: 10.2174/1381612823666170622101319. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

