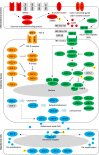
Dauer signalling pathway model for Haemonchus contortus
- PMID: 31036054
- PMCID: PMC6489264
- DOI: 10.1186/s13071-019-3419-6
Dauer signalling pathway model for Haemonchus contortus
Abstract
Background: Signalling pathways have been extensively investigated in the free-living nematode Caenorhabditis elegans, but very little is known about these pathways in parasitic nematodes. Here, we constructed a model for the dauer-associated signalling pathways in an economically highly significant parasitic worm, Haemonchus contortus.
Methods: Guided by data and information available for C. elegans, we used extensive genomic and transcriptomic datasets to infer gene homologues in the dauer-associated pathways, explore developmental transcriptomic, proteomic and phosphoproteomic profiles in H. contortus and study selected molecular structures.
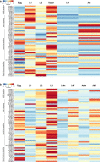
Results: The canonical cyclic guanosine monophosphate (cGMP), transforming growth factor-β (TGF-β), insulin-like growth factor 1 (IGF-1) and steroid hormone signalling pathways of H. contortus were inferred to represent a total of 61 gene homologues. Compared with C. elegans, H. contortus has a reduced set of genes encoding insulin-like peptides, implying evolutionary and biological divergences between the parasitic and free-living nematodes. Similar transcription profiles were found for all gene homologues between the infective stage of H. contortus and dauer stage of C. elegans. High transcriptional levels for genes encoding G protein-coupled receptors (GPCRs), TGF-β, insulin-like ligands (e.g. ins-1, ins-17 and ins-18) and transcriptional factors (e.g. daf-16) in the infective L3 stage of H. contortus were suggestive of critical functional roles in this stage. Conspicuous protein expression patterns and extensive phosphorylation of some components of these pathways suggested marked post-translational modifications also in the L3 stage. The high structural similarity in the DAF-12 ligand binding domain among nematodes indicated functional conservation in steroid (i.e. dafachronic acid) signalling linked to worm development.
Conclusions: Taken together, this pathway model provides a basis to explore hypotheses regarding biological processes and regulatory mechanisms (via particular microRNAs, phosphorylation events and/or lipids) associated with the development of H. contortus and related nematodes as well as parasite-host cross talk, which could aid the discovery of new therapeutic targets.
Keywords: Dauer signalling pathway; Haemonchus contortus; Phospho-proteomic analysis; Proteomic analysis; Transcriptomic analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Structural and functional characterisation of the fork head transcription factor-encoding gene, Hc-daf-16, from the parasitic nematode Haemonchus contortus (Strongylida).Int J Parasitol. 2010 Mar 15;40(4):405-15. doi: 10.1016/j.ijpara.2009.09.005. Epub 2009 Sep 29. Int J Parasitol. 2010. PMID: 19796644 Free PMC article.
-
Identification and characterization of an R-Smad homologue (Hco-DAF-8) from Haemonchus contortus.Parasit Vectors. 2020 Apr 3;13(1):164. doi: 10.1186/s13071-020-04034-0. Parasit Vectors. 2020. PMID: 32245505 Free PMC article.
-
Hc-daf-2 encodes an insulin-like receptor kinase in the barber's pole worm, Haemonchus contortus, and restores partial dauer regulation.Int J Parasitol. 2014 Jun;44(7):485-96. doi: 10.1016/j.ijpara.2014.03.005. Epub 2014 Apr 12. Int J Parasitol. 2014. PMID: 24727120 Free PMC article.
-
Elucidating the molecular and developmental biology of parasitic nematodes: Moving to a multiomics paradigm.Adv Parasitol. 2020;108:175-229. doi: 10.1016/bs.apar.2019.12.005. Epub 2020 Jan 31. Adv Parasitol. 2020. PMID: 32291085 Review.
-
The barber's pole worm CAP protein superfamily--A basis for fundamental discovery and biotechnology advances.Biotechnol Adv. 2015 Dec;33(8):1744-54. doi: 10.1016/j.biotechadv.2015.07.003. Epub 2015 Jul 31. Biotechnol Adv. 2015. PMID: 26239368 Review.
Cited by
-
Research progress and limitation analysis of RNA interference in Haemonchus contortus in China.Front Vet Sci. 2023 Feb 24;10:1079676. doi: 10.3389/fvets.2023.1079676. eCollection 2023. Front Vet Sci. 2023. PMID: 36908509 Free PMC article. Review.
-
Helminth lipidomics: Technical aspects and future prospects.Curr Res Parasitol Vector Borne Dis. 2021 Feb 24;1:100018. doi: 10.1016/j.crpvbd.2021.100018. eCollection 2021. Curr Res Parasitol Vector Borne Dis. 2021. PMID: 35284853 Free PMC article. Review.
-
Dafachronic acid and temperature regulate canonical dauer pathways during Nippostrongylus brasiliensis infectious larvae activation.Parasit Vectors. 2020 Apr 1;13(1):162. doi: 10.1186/s13071-020-04035-z. Parasit Vectors. 2020. PMID: 32238181 Free PMC article.
-
Rhabditophanes diutinus a parthenogenetic clade IV nematode with dauer larvae.PLoS Pathog. 2020 Dec 3;16(12):e1009113. doi: 10.1371/journal.ppat.1009113. eCollection 2020 Dec. PLoS Pathog. 2020. PMID: 33270811 Free PMC article.
-
Dauer fate in a Caenorhabditis elegans Boolean network model.PeerJ. 2023 Jan 23;11:e14713. doi: 10.7717/peerj.14713. eCollection 2023. PeerJ. 2023. PMID: 36710867 Free PMC article.
References
-
- Gushchin I, Gordeliy V. Transmembrane signal transduction in two-component systems: piston, scissoring, or helical rotation? Bioessays. 2018;40. 10.1002/bies201700197. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

