Defining the core essential genome of Pseudomonas aeruginosa
- PMID: 31036669
- PMCID: PMC6525520
- DOI: 10.1073/pnas.1900570116
Defining the core essential genome of Pseudomonas aeruginosa
Abstract
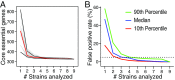
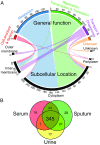
Genomics offered the promise of transforming antibiotic discovery by revealing many new essential genes as good targets, but the results fell short of the promise. While numerous factors contributed to the disappointing yield, one factor was that essential genes for a bacterial species were often defined based on a single or limited number of strains grown under a single or limited number of in vitro laboratory conditions. In fact, the essentiality of a gene can depend on both the genetic background and growth condition. We thus developed a strategy for more rigorously defining the core essential genome of a bacterial species by studying many pathogen strains and growth conditions. We assessed how many strains must be examined to converge on a set of core essential genes for a species. We used transposon insertion sequencing (Tn-Seq) to define essential genes in nine strains of Pseudomonas aeruginosa on five different media and developed a statistical model, FiTnEss, to classify genes as essential versus nonessential across all strain-medium combinations. We defined a set of 321 core essential genes, representing 6.6% of the genome. We determined that analysis of four strains was typically sufficient in P. aeruginosa to converge on a set of core essential genes likely to be essential across the species across a wide range of conditions relevant to in vivo infection, and thus to represent attractive targets for novel drug discovery.
Keywords: ESKAPE; Tn-Seq; antibiotic discovery; gram-negative pathogens.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: Eric Lander serves on the Board of Directors for and holds equity in Codiak BioSciences and Neon Therapeutics, and serves on the Scientific Advisory Board of F-Prime Capital Partners and Third Rock Ventures; he is also affiliated with several nonprofit organizations, including serving on the Board of Directors of the Innocence Project, Count Me In, and Biden Cancer Initiative, and the Board of Trustees for the Parker Institute for Cancer Immunotherapy. He has served and continues to serve on various federal advisory committees.
Figures



References
-
- Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. - PubMed
-
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. - PubMed
-
- Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015;14:529–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

