Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA)
- PMID: 31037156
- PMCID: PMC6485294
- DOI: 10.7150/thno.28119
Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA)
Abstract
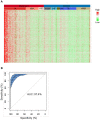
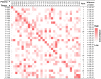
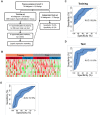
Rational: LDCT screening can identify early-stage lung cancers yet introduces excessive false positives and it remains a great challenge to differentiate malignant tumors from benign solitary pulmonary nodules, which calls for better non-invasive diagnostic tools. Methods: We performed DNA methylation profiling by high throughput DNA bisulfite sequencing in tissue samples (nodule size < 3 cm in diameter) to learn methylation patterns that differentiate cancerous tumors from benign lesions. Then we filtered out methylation patterns exhibiting high background in circulating tumor DNA (ctDNA) and built an assay for plasma sample classification. Results: We first performed methylation profiling of 230 tissue samples to learn cancer-specific methylation patterns which achieved a sensitivity of 92.7% (88.3% - 97.1%) and a specificity of 92.8% (89.3% - 96.3%). These tissue-derived DNA methylation markers were further filtered using a training set of 66 plasma samples and 9 markers were selected to build a diagnostic prediction model. From an independent validation set of additional 66 plasma samples, this model obtained a sensitivity of 79.5% (63.5% - 90.7%) and a specificity of 85.2% (66.3% - 95.8%) for differentiating patients with malignant tumor (n = 39) from patients with benign lesions (n = 27). Additionally, when tested on gender and age matched asymptomatic normal individuals (n = 118), our model achieved a specificity of 93.2% (89.0% - 98.3%). Specifically, our assay is highly sensitive towards early-stage lung cancer, with a sensitivity of 75.0% (55.0%-90.0%) in 20 stage Ia lung cancer patients and 85.7% (57.1%-100.0%) in 7 stage Ib lung cancer patients. Conclusions: We have developed a novel sensitive blood based non-invasive diagnostic assay for detecting early stage lung cancer as well as differentiating lung cancers from benign pulmonary nodules.
Keywords: Early-stage lung cancer; circulating tumor DNA; high-throughput targeted DNA methylation sequencing.
Conflict of interest statement
Competing Interests: The authors JBF, XC, YG, MY, WX, YZ, JT, and ZC are employees of AnchorDx Medical Co., Ltd., a company that focuses on the development of next generation sequencing diagnostic products for early cancer detection using liquid biopsy. The author PWL is a member of AnchorDx's Scientific Advisory Board. All other authors declare no competing financial interest.
Figures

References
-
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. - PubMed
-
- Li S, Zhao B, Wang X, Yu J, Yan S, Lv C. et al. Overestimated value of (18)F-FDG PET/CT to diagnose pulmonary nodules: Analysis of 298 patients. Clin Radiol. 2014;69:e352–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

