G1/S cell cycle regulators mediate effects of circadian dysregulation on tumor growth and provide targets for timed anticancer treatment
- PMID: 31039152
- PMCID: PMC6490878
- DOI: 10.1371/journal.pbio.3000228
G1/S cell cycle regulators mediate effects of circadian dysregulation on tumor growth and provide targets for timed anticancer treatment
Abstract
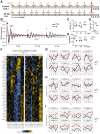
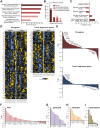
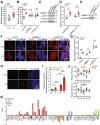
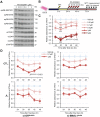
Circadian disruption has multiple pathological consequences, but the underlying mechanisms are largely unknown. To address such mechanisms, we subjected transformed cultured cells to chronic circadian desynchrony (CCD), mimicking a chronic jet-lag scheme, and assayed a range of cellular functions. The results indicated a specific circadian clock-dependent increase in cell proliferation. Transcriptome analysis revealed up-regulation of G1/S phase transition genes (myelocytomatosis oncogene cellular homolog [Myc], cyclin D1/3, chromatin licensing and DNA replication factor 1 [Cdt1]), concomitant with increased phosphorylation of the retinoblastoma (RB) protein by cyclin-dependent kinase (CDK) 4/6 and increased G1-S progression. Phospho-RB (Ser807/811) was found to oscillate in a circadian fashion and exhibit phase-shifted rhythms in circadian desynchronized cells. Consistent with circadian regulation, a CDK4/6 inhibitor approved for cancer treatment reduced growth of cultured cells and mouse tumors in a time-of-day-specific manner. Our study identifies a mechanism that underlies effects of circadian disruption on tumor growth and underscores the use of treatment timed to endogenous circadian rhythms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

