Whole-genome and targeted sequencing of drug-resistant Mycobacterium tuberculosis on the iSeq100 and MiSeq: A performance, ease-of-use, and cost evaluation
- PMID: 31039166
- PMCID: PMC6490892
- DOI: 10.1371/journal.pmed.1002794
Whole-genome and targeted sequencing of drug-resistant Mycobacterium tuberculosis on the iSeq100 and MiSeq: A performance, ease-of-use, and cost evaluation
Erratum in
-
Correction: Whole-genome and targeted sequencing of drug-resistant Mycobacterium tuberculosis on the iSeq100 and MiSeq: A performance, ease-of-use, and cost evaluation.PLoS Med. 2019 Jun 3;16(6):e1002823. doi: 10.1371/journal.pmed.1002823. eCollection 2019 Jun. PLoS Med. 2019. PMID: 31158219 Free PMC article.
Abstract
Background: Accurate, comprehensive, and timely detection of drug-resistant tuberculosis (TB) is essential to inform patient treatment and enable public health surveillance. This is crucial for effective control of TB globally. Whole-genome sequencing (WGS) and targeted next-generation sequencing (NGS) approaches have potential as rapid in vitro diagnostics (IVDs), but the complexity of workflows, interpretation of results, high costs, and vulnerability of instrumentation have been barriers to broad uptake outside of reference laboratories, especially in low- and middle-income countries. A new, solid-state, tabletop sequencing instrument, Illumina iSeq100, has the potential to decentralize NGS for individual patient care.
Methods and findings: In this study, we evaluated WGS and targeted NGS for TB on both the new iSeq100 and the widely used MiSeq (both manufactured by Illumina) and compared sequencing performance, costs, and usability. We utilized DNA libraries produced from Mycobacterium tuberculosis clinical isolates for the evaluation. We conducted WGS on three strains and observed equivalent uniform genome coverage with both platforms and found the depth of coverage obtained was consistent with the expected data output. Utilizing the standardized, cloud-based ReSeqTB bioinformatics pipeline for variant analysis, we found the two platforms to have 94.0% (CI 93.1%-94.8%) agreement, in comparison to 97.6% (CI 97%-98.1%) agreement for the same libraries on two MiSeq instruments. For the targeted NGS approach, 46 M. tuberculosis-specific amplicon libraries had 99.6% (CI 98.0%-99.9%) agreement between the iSeq100 and MiSeq data sets in drug resistance-associated SNPs. The upfront capital costs are almost 5-fold lower for the iSeq100 ($19,900 USD) platform in comparison to the MiSeq ($99,000 USD); however, because of difference in the batching capabilities, the price per sample for WGS was higher on the iSeq100. For WGS of M. tuberculosis at the minimum depth of coverage of 30x, the cost per sample on the iSeq100 was $69.44 USD versus $28.21 USD on the MiSeq, assuming a 2 × 150 bp run on a v3 kit. In terms of ease of use, the sequencing workflow of iSeq100 has been optimized to only require 27 minutes total of hands-on time pre- and post-run, and the maintenance is simplified by a single-use cartridge-based fluidic system. As these are the first sequencing attempts on the iSeq100 for M. tuberculosis, the sequencing pool loading concentration still needs optimization, which will affect sequencing error and depth of coverage. Additionally, the costs are based on current equipment and reagent costs, which are subject to change.
Conclusions: The iSeq100 instrument is capable of running existing TB WGS and targeted NGS library preparations with comparable accuracy to the MiSeq. The iSeq100 has reduced sequencing workflow hands-on time and is able to deliver sequencing results in <24 hours. Reduced capital and maintenance costs and lower-throughput capabilities also give the iSeq100 an advantage over MiSeq in settings of individualized care but not in high-throughput settings such as reference laboratories, where sample batching can be optimized to minimize cost at the expense of workflow complexity and time.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: DL and DME work for the nonprofit Translational Genomics Research Institute (TGen), which holds the patent on the Next Gen RDST assay that was employed in this study. As of this submission, TGen has not licensed the assay to any commercial party. JH is currently a paid employee of Illumina, where he works as a scientist. AGY is currently an employee of Illumina and a shareholder. CMD served as a Guest Editor on PLOS Medicine’s Special Issue on Tuberculosis.
Figures
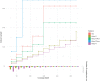
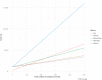
References
-
- World; Health Organization (WHO). Global Tuberculosis Report 2018. Geneva, Switzerland: World Health Organization (WHO); 2018.
-
- (WHO) WHO. Implementing the End TB Strategy: the essentials; 2015. Report No.: WHO/HTM/TB/2015.31.
-
- Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. The European respiratory journal. 2017;50(6). 10.1183/13993003.01354-2017 erj.ersjournals.com. - DOI - PMC - PubMed
-
- (WHO) WHO. The use of next-generation sequencing technologies for the detection of mutations associated with drug resistance in Mycobacterium tuberculosis complex: technical guide; 2018. Report No.: WHO/CDS/TB/2018.19.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

