Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma
- PMID: 31039795
- PMCID: PMC6492406
- DOI: 10.1186/s13073-019-0636-8
Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma
Abstract
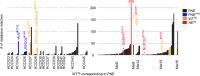
Background: Although mutated HLA ligands are considered ideal cancer-specific immunotherapy targets, evidence for their presentation is lacking in hepatocellular carcinomas (HCCs). Employing a unique multi-omics approach comprising a neoepitope identification pipeline, we assessed exome-derived mutations naturally presented as HLA class I ligands in HCCs.
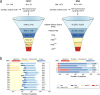
Methods: In-depth multi-omics analyses included whole exome and transcriptome sequencing to define individual patient-specific search spaces of neoepitope candidates. Evidence for the natural presentation of mutated HLA ligands was investigated through an in silico pipeline integrating proteome and HLA ligandome profiling data.
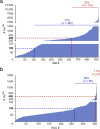
Results: The approach was successfully validated in a state-of-the-art dataset from malignant melanoma, and despite multi-omics evidence for somatic mutations, mutated naturally presented HLA ligands remained elusive in HCCs. An analysis of extensive cancer datasets confirmed fundamental differences of tumor mutational burden in HCC and malignant melanoma, challenging the notion that exome-derived mutations contribute relevantly to the expectable neoepitope pool in malignancies with only few mutations.
Conclusions: This study suggests that exome-derived mutated HLA ligands appear to be rarely presented in HCCs, inter alia resulting from a low mutational burden as compared to other malignancies such as malignant melanoma. Our results therefore demand widening the target scope for personalized immunotherapy beyond this limited range of mutated neoepitopes, particularly for malignancies with similar or lower mutational burden.
Keywords: HLA; HLA ligandomics; Hepatocellular carcinoma; Immunoinformatics; Immunotherapy; Liver cancer; Mass spectrometry; Multi-omics; Neoantigen; Next-generation sequencing; Peptide prediction; Personalized medicine.
Conflict of interest statement
Ethics approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki and applicable laws and regulations and has been approved by the local institutional review board at the University Hospital of Tübingen, Germany (Project No. 364/2014BO2). All participants provided written informed consent before study inclusion.
Consent for publication
Not applicable.
Competing interests
M.W. Löffler, D.J. Kowalewski, H. Schuster, S. Stevanović, and S.P. Haen are the inventors of patents owned by Immatics Biotechnologies GmbH. D.J. Kowalewski and H. Schuster are currently employees of Immatics Biotechnologies GmbH. H.G. Rammensee has ownership interest (including patents) in Immatics Biotechnologies GmbH, CureVac AG, and Synimmune GmbH. The remaining authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- European Association for the Study of the Liver, European Organisation for Research and Treatment of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56(4):908–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

