Epigenome-wide association study for lifetime estrogen exposure identifies an epigenetic signature associated with breast cancer risk
- PMID: 31039828
- PMCID: PMC6492393
- DOI: 10.1186/s13148-019-0664-7
Epigenome-wide association study for lifetime estrogen exposure identifies an epigenetic signature associated with breast cancer risk
Abstract
Background: It is well established that estrogens and other hormonal factors influence breast cancer susceptibility. We hypothesized that a woman's total lifetime estrogen exposure accumulates changes in DNA methylation, detectable in the blood, which could be used in risk assessment for breast cancer.
Methods: An estimated lifetime estrogen exposure (ELEE) model was defined using epidemiological data from EPIC-Italy (n = 31,864). An epigenome-wide association study (EWAS) of ELEE was performed using existing Illumina HumanMethylation450K Beadchip (HM450K) methylation data obtained from EPIC-Italy blood DNA samples (n = 216). A methylation index (MI) of ELEE based on 31 CpG sites was developed using HM450K data from EPIC-Italy and the Generations Study and evaluated for association with breast cancer risk in an independent dataset from the Generations Study (n = 440 incident breast cancer cases matched to 440 healthy controls) using targeted bisulfite sequencing. Lastly, a meta-analysis was conducted including three additional cohorts, consisting of 1187 case-control pairs.
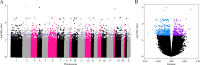
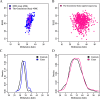
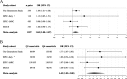
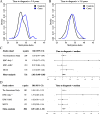
Results: We observed an estimated 5% increase in breast cancer risk per 1-year longer ELEE (OR = 1.05, 95% CI 1.04-1.07, P = 3 × 10-12) in EPIC-Italy. The EWAS identified 694 CpG sites associated with ELEE (FDR Q < 0.05). We report a DNA methylation index (MI) associated with breast cancer risk that is validated in the Generations Study targeted bisulfite sequencing data (ORQ4_vs_Q1 = 1.77, 95% CI 1.07-2.93, P = 0.027) and in the meta-analysis (ORQ4_vs_Q1 = 1.43, 95% CI 1.05-2.00, P = 0.024); however, the correlation between the MI and ELEE was not validated across study cohorts.
Conclusion: We have identified a blood DNA methylation signature associated with breast cancer risk in this study. Further investigation is required to confirm the interaction between estrogen exposure and DNA methylation in the blood.
Keywords: Biomarker; Breast cancer; Cancer risk; DNA methylation; EWAS; Epigenetics; Estrogen exposure; Hormonal exposures.
Conflict of interest statement
Ethics approval and consent to participate
All study participants signed informed consent forms, and each cohort was approved by the national ethical review boards: The Generations Study: South Thames Multicentre Research Ethics Committee (reference MREC 03/01/014), and EPIC-Italy and EPIC-IARC: The ethics committees of the Human Genetics Foundation (HuGeF) and IARC respectively. MCCS: The MCCS study protocol was approved by the Cancer Council Victoria’s Human Research Ethics.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- International Agency for Research on Cancer. EPIC study. 2018. http://epic.iarc.fr. Accessed: September 2018.
-
- Cancer Research UK. Breast cancer statistics 2018. https://www.cancerresearchuk.org/health-professional/cancer-statistics/s.... Accessed: November 2018.
-
- Endogenous Hormones Breast Cancer Collaborative Group Sex hormones and risk of breast cancer in premenopausal women: a collaborative reanalysis of individual participant data from seven prospective studies. Lancet Oncol. 2013;14(10):1009–1019. doi: 10.1016/S1470-2045(13)70301-2. - DOI - PMC - PubMed

