Development of a novel cultivation technique for uncultured soil bacteria
- PMID: 31040339
- PMCID: PMC6491550
- DOI: 10.1038/s41598-019-43182-x
Development of a novel cultivation technique for uncultured soil bacteria
Abstract
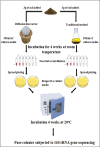
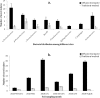
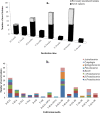
In this study, a new diffusion bioreactor was developed to cultivate hidden bacterial communities in their natural environment. The newly developed method was investigated to cultivate microbial communities from the forest soil, and the results were evaluated against traditional culture methods and compared to the results of a pyrosequencing-based molecular survey. The molecular analysis revealed that a diverse bacterial population was present in the soil sample. However, both the newly developed method and the traditional method recovered more than 400 isolates, which belonged to only four phyla: Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes. Although these isolates were distributed over only four major phyla, the use of the newly developed technique resulted in the successful cultivation of 35 previously uncultured strains, whereas no such strains were successfully cultivated by the traditional method. Furthermore, the study also found that the recovery of uncultured bacteria and novel isolates was related to sampling season, incubation period, and cultivation media. The use of soil collected in summer, a prolonged incubation period, and low-substrate modified media increased the recovery of uncultured and novel isolates. Overall, the results indicate that the newly designed diffusion bioreactor can mimic the natural environment, which permits the cultivation of previously uncultured bacteria.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

