lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway
- PMID: 31040703
- PMCID: PMC6462170
- DOI: 10.2147/OTT.S188054
lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway
Abstract
Purpose: The chemoresistance of colon cancer to oxaliplatin (L-OHP) indicates poor prognosis. Long non-coding RNA (lncRNA) KCNQ1OT1 (KCNQ1 opposite strand/antisense transcript 1) has been shown to participate in the tumorigenesis of several types of cancers. However, little is known about the role of KCNQ1OT1 in the chemoresistance and prognosis of colon cancer.
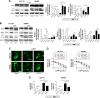
Materials and methods: Quantitative-PCR and Western blot were used to measure the expression profiles of KCNQ1OT1, miR-34a, and Atg4B in colon cancer tissues and cells. Cell viability assay and flow cytometry were used to examine their effects on cell proliferation and death. Cleavage of LC3 and GFP-LC3 plasmid transfection were used to detect autophagic activity. Double luciferase reporter assay was used to verify the interactions between miRNA and lncRNA or mRNA. Xenograft tumor model was used to verify the effects of KCNQ1OT1 in vivo.
Results: In this study, it is shown that the expression level of KCNQ1OT1 was increased in tumor, which indicated poor prognosis in colon cancer patients. Using colon cancer cell lines HCT116 and SW480, it was demonstrated that knockdown of KCNQ1OT1 decreased the cell viability and increased the apoptosis rates upon L-OHP treatment. Further studies indicated that Atg4B upregulation was partially responsible for KCNQ1OT1-induced protective autophagy and chemoresistance. Moreover, miR-34a functioned as a bridge between KCNQ1OT1 and Atg4B, which could be sponged by KCNQ1OT1, while it could also bind to the 3'-UTR of Atg4B and downregulate its expressions. Finally, we show that the KCNQ1OT1/miR-34a/Atg4B axis regulated the chemoresistance of colon cancer cells in vitro and in vivo.
Conclusion: lncRNA KCNQ1OT1 promoted the chemoresistance of colon cancer by sponging miR-34a, thus upregulating the expressions of Atg4B and enhancing protective autophagy. KCNQ1OT1 might become a promising target for colon cancer therapeutics.
Keywords: Atg4B; KCNQ1OT1; chemoresistance; colorectal cancer; miR-34a; protective autophagy.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures



References
LinkOut - more resources
Full Text Sources

