Understanding the Contributions of Conformational Changes, Thermodynamics, and Kinetics of RNA-Small Molecule Interactions
- PMID: 31042354
- PMCID: PMC6525067
- DOI: 10.1021/acschembio.8b00945
Understanding the Contributions of Conformational Changes, Thermodynamics, and Kinetics of RNA-Small Molecule Interactions
Abstract
The implication of RNA in multiple cellular processes beyond protein coding has revitalized interest in the development of small molecules for therapeutically targeting RNA and for further probing its cellular biology. However, the process of rationally designing such small molecule probes is hampered by the paucity of information about fundamental molecular recognition principles of RNA. In this Review, we summarize two important and often underappreciated aspects of RNA-small molecule recognition: RNA conformational dynamics and the biophysical properties of interactions of small molecules with RNA, specifically thermodynamics and kinetics. While conformational flexibility is often said to impede RNA ligand development, the ability of small molecules to influence the RNA conformational landscape can have a significant effect on the cellular functions of RNA. An analysis of the conformational landscape of RNA and the interactions of individual conformations with ligands can thus guide the development of new small molecule probes, which needs to be investigated further. Additionally, while it is common practice to quantify the binding affinities ( Ka or Kd) of small molecules for biomacromolecules as a measure of their activity, further biophysical characterization of their interaction can provide a deeper understanding. Studies that focus on the thermodynamic and kinetic parameters for interaction between RNA and ligands are next discussed. Finally, this Review provides the reader with a perspective on how such in-depth analysis of biophysical characteristics of the interaction of RNA and small molecules can impact our understanding of these interactions and how they will benefit the future design of small molecule probes.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
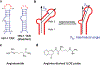
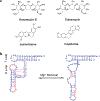
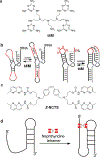
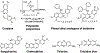
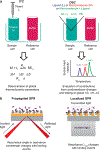

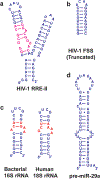
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

