Single-cell transcriptomic analysis of Alzheimer's disease
- PMID: 31042697
- PMCID: PMC6865822
- DOI: 10.1038/s41586-019-1195-2
Single-cell transcriptomic analysis of Alzheimer's disease
Erratum in
-
Author Correction: Single-cell transcriptomic analysis of Alzheimer's disease.Nature. 2019 Jul;571(7763):E1. doi: 10.1038/s41586-019-1329-6. Nature. 2019. PMID: 31209304 Free PMC article.
Abstract
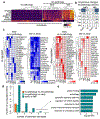
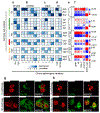
Alzheimer's disease is a pervasive neurodegenerative disorder, the molecular complexity of which remains poorly understood. Here, we analysed 80,660 single-nucleus transcriptomes from the prefrontal cortex of 48 individuals with varying degrees of Alzheimer's disease pathology. Across six major brain cell types, we identified transcriptionally distinct subpopulations, including those associated with pathology and characterized by regulators of myelination, inflammation, and neuron survival. The strongest disease-associated changes appeared early in pathological progression and were highly cell-type specific, whereas genes upregulated at late stages were common across cell types and primarily involved in the global stress response. Notably, we found that female cells were overrepresented in disease-associated subpopulations, and that transcriptional responses were substantially different between sexes in several cell types, including oligodendrocytes. Overall, myelination-related processes were recurrently perturbed in multiple cell types, suggesting that myelination has a key role in Alzheimer's disease pathophysiology. Our single-cell transcriptomic resource provides a blueprint for interrogating the molecular and cellular basis of Alzheimer's disease.
Conflict of interest statement
Figures











Comment in
-
An 80,000-Piece Puzzle of Alzheimer's Disease.Immunity. 2019 Jun 18;50(6):1349-1351. doi: 10.1016/j.immuni.2019.05.016. Immunity. 2019. PMID: 31216459
References
-
- Masters CL et al. Alzheimer’s disease. Nat Rev Dis Primers 1, 15056 (2015). - PubMed
-
- Hardy J & Selkoe DJ The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002). - PubMed
-
- Braak H & Braak E Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991). - PubMed
-
- De Strooper B & Karran E The Cellular Phase of Alzheimer’s Disease. Cell 164, 603–615 (2016). - PubMed
-
- Canter RG, Penney J & Tsai L-H The road to restoring neural circuits for the treatment of Alzheimer’s disease. Nature 539, 187–196 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 AG062421/AG/NIA NIH HHS/United States
- RF1 AG062377/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- RF1 AG057473/AG/NIA NIH HHS/United States
- RF1 AG054321/AG/NIA NIH HHS/United States
- U01 NS110453/NS/NINDS NIH HHS/United States
- R01 AG062335/AG/NIA NIH HHS/United States
- R01 MH109978/MH/NIMH NIH HHS/United States
- U01 MH119509/MH/NIMH NIH HHS/United States
- RF1 AG054012/AG/NIA NIH HHS/United States
- U01 AG046152/AG/NIA NIH HHS/United States
- R01 AG058002/AG/NIA NIH HHS/United States
- R01 HG008155/HG/NHGRI NIH HHS/United States
- K08 EY026652/EY/NEI NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- R01 AG046174/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

