Performance of Three Common Hepatitis C Virus (HCV) Genotyping Assays for Identification of HCV Genotype 2/1 Chimeras
- PMID: 31043467
- PMCID: PMC6595458
- DOI: 10.1128/JCM.00060-19
Performance of Three Common Hepatitis C Virus (HCV) Genotyping Assays for Identification of HCV Genotype 2/1 Chimeras
Abstract
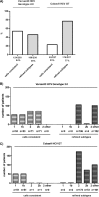
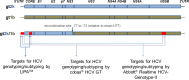
Besides seven major hepatitis C virus (HCV) genotypes (GT), a number of intergenotypic recombinant strains have been described. These so-called chimeras combine genetic characteristics of different HCV genotypes. However, correct genotype classification is important, as choice and duration of direct-acting antiviral (DAA) treatment is mainly based on the viral genotype. Therefore, misclassification of chimeras might lead to suboptimal treatment of patients infected with these strains. For example, 2k/1b chimeras are typically described as HCV genotype 2 strains by commercially available hybridization assays, but real-time PCR-based tests recognizing another HCV region might be more suitable for correct chimera detection. In this study, the analytic capacity of the hybridization-assay Versant HCV Genotype 2.0 (LiPA 2.0) and the real-time PCR-based-assays cobas HCV GT and Abbott RealTime HCV Genotype II were tested in a selected cohort of 230 patients infected with HCV genotype 1 (n = 53) and 2 (n = 177) and 48 patients infected with HCV 2/1 chimeric strains. While the Versant HCV Genotype 2.0 (LiPA 2.0) assay failed to identify chimeras in all of the patients (48/48, 100%), cobas HCV GT and Abbott HCV Genotype II assays identified chimeras correctly in 90% (43/48) and 65% (31/48) of the cases, respectively. In conclusion, while the hybridization-based Versant HCV Genotype 2.0 (LiPA 2.0) assay seems to be unsuitable for detection of HCV 2/1 chimeras, use of the real-time PCR-based assays cobas HCV GT and Abbott RealTime HCV Genotype II led to a higher rate of chimera detection.
Keywords: 2k/1b; assay; chimera; hepatitis C virus; recombinant.
Copyright © 2019 American Society for Microbiology.
Figures

References
-
- Hedskog C, Doehle B, Chodavarapu K, Gontcharova V, Crespo Garcia J, De Knegt R, Drenth JP, McHutchison JG, Brainard D, Stamm LM, Miller MD, Svarovskaia E, Mo H. 2015. Characterization of hepatitis C virus intergenotypic recombinant strains and associated virological response to sofosbuvir/ribavirin. Hepatology 61:471–480. doi:10.1002/hep.27361. - DOI - PubMed
-
- Susser S, Dietz J, Schlevogt B, Zuckerman E, Barak M, Piazzolla V, Howe A, Hinrichsen H, Passmann S, Daniel R, Cornberg M, Mangia A, Zeuzem S, Sarrazin C. 2017. Origin, prevalence and response to therapy of hepatitis C virus genotype 2k/1b chimeras. J Hepatol 67:680–686. doi:10.1016/j.jhep.2017.05.027. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

