Human Monoclonal Antibodies Potently Neutralize Zika Virus and Select for Escape Mutations on the Lateral Ridge of the Envelope Protein
- PMID: 31043537
- PMCID: PMC6600209
- DOI: 10.1128/JVI.00405-19
Human Monoclonal Antibodies Potently Neutralize Zika Virus and Select for Escape Mutations on the Lateral Ridge of the Envelope Protein
Abstract
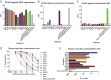
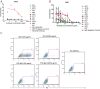
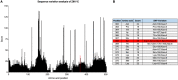
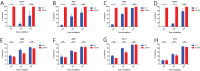
The mosquito-borne Zika virus (ZIKV) has been causing epidemic outbreaks on a global scale. Virus infection can result in severe disease in humans, including microcephaly in newborns and Guillain-Barré syndrome in adults. Here, we characterized monoclonal antibodies isolated from a patient with an active Zika virus infection that potently neutralized virus infection in Vero cells at the nanogram-per-milliliter range. In addition, these antibodies enhanced internalization of virions into human leukemia K562 cells in vitro, indicating their possible ability to cause antibody-dependent enhancement of disease. Escape variants of the ZIKV MR766 strain to a potently neutralizing antibody, AC10, exhibited an amino acid substitution at residue S368 in the lateral ridge region of the envelope protein. Analysis of publicly availably ZIKV sequences revealed the S368 site to be conserved among the vast majority (97.6%) of circulating strains. We validated the importance of this residue by engineering a recombinant virus with an S368R point mutation that was unable to be fully neutralized by AC10. Four out of the 12 monoclonal antibodies tested were also unable to neutralize the virus with the S368R mutation, suggesting this region to be an important immunogenic epitope during human infection. Last, a time-of-addition infection assay further validated the escape variant and showed that all monoclonal antibodies inhibited virus binding to the cell surface. Thus, the present study demonstrates that the lateral ridge region of the envelope protein is likely an immunodominant, neutralizing epitope.IMPORTANCE Zika virus (ZIKV) is a global health threat causing severe disease in humans, including microcephaly in newborns and Guillain-Barré syndrome in adults. Here, we analyzed the human monoclonal antibody response to acute ZIKV infection and found that neutralizing antibodies could not elicit Fc-mediated immune effector functions but could potentiate antibody-dependent enhancement of disease. We further identified critical epitopes involved with neutralization by generating and characterizing escape variants by whole-genome sequencing. We demonstrate that the lateral ridge region, particularly the S368 amino acid site, is critical for neutralization by domain III-specific antibodies.
Keywords: Zika virus; antibody-dependent cellular cytotoxicity; monoclonal antibodies; neutralizing antibodies; prM-E; whole-genome sequencing.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- Cao-Lormeau V-M, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial A-L, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra J-C, Despres P, Fournier E, Mallet H-P, Musso D, Fontanet A, Neil J, Ghawché F. 2016. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387:1531–1539. doi:10.1016/S0140-6736(16)00562-6. - DOI - PMC - PubMed
-
- Brasil P, Pereira JP, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai U-A, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Raja Gabaglia C, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baião AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. 2016. Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med 375:2321–2334. doi:10.1056/NEJMoa1602412. - DOI - PMC - PubMed
-
- Gaudinski MR, Houser KV, Morabito KM, Hu Z, Yamshchikov G, Rothwell RS, Berkowitz N, Mendoza F, Saunders JG, Novik L, Hendel CS, Holman LA, Gordon IJ, Cox JH, Edupuganti S, McArthur MA, Rouphael NG, Lyke KE, Cummings GE, Sitar S, Bailer RT, Foreman BM, Burgomaster K, Pelc RS, Gordon DN, DeMaso CR, Dowd KA, Laurencot C, Schwartz RM, Mascola JR, Graham BS, Pierson TC, Ledgerwood JE, Chen GL, Plummer S, Costner P, Zephir K, Casazza J, Ola A, Victorino M, Levinson C, Whalen W, Wang X, Cunningham J, Vasilenko O, Burgos Florez M, Hickman S, Pittman I, Le L, Larkin B, et al. 2018. Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: randomised, open-label, phase 1 clinical trials. Lancet 391:552–562. doi:10.1016/S0140-6736(17)33105-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

