CXCL4 assembles DNA into liquid crystalline complexes to amplify TLR9-mediated interferon-α production in systemic sclerosis
- PMID: 31043596
- PMCID: PMC6494823
- DOI: 10.1038/s41467-019-09683-z
CXCL4 assembles DNA into liquid crystalline complexes to amplify TLR9-mediated interferon-α production in systemic sclerosis
Abstract
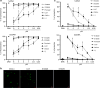
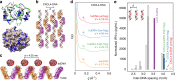
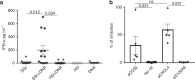
Systemic sclerosis (SSc) is a chronic autoimmune disease characterized by fibrosis and vasculopathy. CXCL4 represents an early serum biomarker of severe SSc and likely contributes to inflammation via chemokine signaling pathways, but the exact role of CXCL4 in SSc pathogenesis is unclear. Here, we elucidate an unanticipated mechanism for CXCL4-mediated immune amplification in SSc, in which CXCL4 organizes "self" and microbial DNA into liquid crystalline immune complexes that amplify TLR9-mediated plasmacytoid dendritic cell (pDC)-hyperactivation and interferon-α production. Surprisingly, this activity does not require CXCR3, the CXCL4 receptor. Importantly, we find that CXCL4-DNA complexes are present in vivo and correlate with type I interferon (IFN-I) in SSc blood, and that CXCL4-positive skin pDCs coexpress IFN-I-related genes. Thus, we establish a direct link between CXCL4 overexpression and the IFN-I-gene signature in SSc and outline a paradigm in which chemokines can drastically modulate innate immune receptors without being direct agonists.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

