First Community-Wide, Comparative Cross-Linking Mass Spectrometry Study
- PMID: 31045356
- PMCID: PMC6625963
- DOI: 10.1021/acs.analchem.9b00658
First Community-Wide, Comparative Cross-Linking Mass Spectrometry Study
Abstract
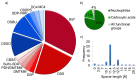
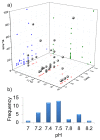
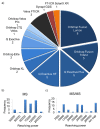
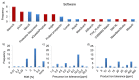
The number of publications in the field of chemical cross-linking combined with mass spectrometry (XL-MS) to derive constraints for protein three-dimensional structure modeling and to probe protein-protein interactions has increased during the last years. As the technique is now becoming routine for in vitro and in vivo applications in proteomics and structural biology there is a pressing need to define protocols as well as data analysis and reporting formats. Such consensus formats should become accepted in the field and be shown to lead to reproducible results. This first, community-based harmonization study on XL-MS is based on the results of 32 groups participating worldwide. The aim of this paper is to summarize the status quo of XL-MS and to compare and evaluate existing cross-linking strategies. Our study therefore builds the framework for establishing best practice guidelines to conduct cross-linking experiments, perform data analysis, and define reporting formats with the ultimate goal of assisting scientists to generate accurate and reproducible XL-MS results.
Conflict of interest statement
The authors declare no competing financial interest.
Figures