An Emergent Integrated Aging Process Conserved Across Primates
- PMID: 31046108
- PMCID: PMC6777080
- DOI: 10.1093/gerona/glz110
An Emergent Integrated Aging Process Conserved Across Primates
Abstract
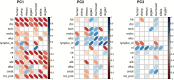
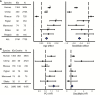
Aging is a complex process emerging from integrated physiological networks. Recent work using principal component analysis (PCA) of multisystem biomarkers proposed a novel fundamental physiological process, "integrated albunemia," which was consistent across human populations and more strongly associated with age and mortality risk than individual biomarkers. Here we tested for integrated albunemia and associations with age and mortality across six diverse nonhuman primate species and humans. PCA of 13 physiological biomarkers recovered in all species a primary axis of variation (PC1) resembling integrated albunemia, which increased with age in all but one species but was less predictive of mortality risk. Within species, PC1 scores were often reliably recovered with a minimal biomarker subset and usually stable between sexes. Even among species, correlations in PC1 structure were often strong, but the effect of phylogeny was inconclusive. Thus, integrated albunemia likely reflects an evolutionarily conserved process across primates and appears to be generally associated with aging but not necessarily with negative impacts on survival. Integrated albunemia is unlikely to be the only conserved emergent physiological process; our findings hence have implications both for the evolution of the aging process and of physiological networks more generally.
Keywords: Biomarkers; Mortality risk; Nonhuman models; Physiological networks; Principal component analysis.
© The Author(s) 2019. Published by Oxford University Press on behalf of The Gerontological Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Han J-DJ, Hou L, Sun N, Xu C, McDermott J, Wang D. The system capacity view of aging and longevity. Quant Biol. 2017;5(3):251–259. doi:10.1007/s40484-017-0115-4 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

