Cross-Regulation between TDP-43 and Paraspeckles Promotes Pluripotency-Differentiation Transition
- PMID: 31047794
- PMCID: PMC6561722
- DOI: 10.1016/j.molcel.2019.03.041
Cross-Regulation between TDP-43 and Paraspeckles Promotes Pluripotency-Differentiation Transition
Abstract
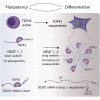
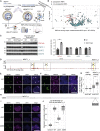
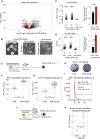
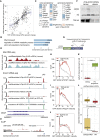
RNA-binding proteins (RBPs) and long non-coding RNAs (lncRNAs) are key regulators of gene expression, but their joint functions in coordinating cell fate decisions are poorly understood. Here we show that the expression and activity of the RBP TDP-43 and the long isoform of the lncRNA Neat1, the scaffold of the nuclear compartment "paraspeckles," are reciprocal in pluripotent and differentiated cells because of their cross-regulation. In pluripotent cells, TDP-43 represses the formation of paraspeckles by enhancing the polyadenylated short isoform of Neat1. TDP-43 also promotes pluripotency by regulating alternative polyadenylation of transcripts encoding pluripotency factors, including Sox2, which partially protects its 3' UTR from miR-21-mediated degradation. Conversely, paraspeckles sequester TDP-43 and other RBPs from mRNAs and promote exit from pluripotency and embryonic patterning in the mouse. We demonstrate that cross-regulation between TDP-43 and Neat1 is essential for their efficient regulation of a broad network of genes and, therefore, of pluripotency and differentiation.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Adriaens C., Standaert L., Barra J., Latil M., Verfaillie A., Kalev P., Boeckx B., Wijnhoven P.W.G., Radaelli E., Vermi W. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016;22:861–868. - PubMed
-
- Alon U. Third Edition. Chapman and Hall/CRC; 2007. An Introduction to Systems Biology: Design Principles of Biological Circuits.
-
- Baltz A.G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

