Mechanism of cognate sequence discrimination by the ETS-family transcription factor ETS-1
- PMID: 31048376
- PMCID: PMC6597803
- DOI: 10.1074/jbc.RA119.007866
Mechanism of cognate sequence discrimination by the ETS-family transcription factor ETS-1
Abstract
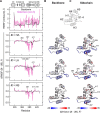
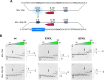
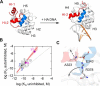
Functional evidence increasingly implicates low-affinity DNA recognition by transcription factors as a general mechanism for the spatiotemporal control of developmental genes. Although the DNA sequence requirements for affinity are well-defined, the dynamic mechanisms that execute cognate recognition are much less resolved. To address this gap, here we examined ETS1, a paradigm developmental transcription factor, as a model for which cognate discrimination remains enigmatic. Using molecular dynamics simulations, we interrogated the DNA-binding domain of murine ETS1 alone and when bound to high-and low-affinity cognate sites or to nonspecific DNA. The results of our analyses revealed collective backbone and side-chain motions that distinguished cognate versus nonspecific as well as high- versus low-affinity cognate DNA binding. Combined with binding experiments with site-directed ETS1 mutants, the molecular dynamics data disclosed a triad of residues that respond specifically to low-affinity cognate DNA. We found that a DNA-contacting residue (Gln-336) specifically recognizes low-affinity DNA and triggers the loss of a distal salt bridge (Glu-343/Arg-378) via a large side-chain motion that compromises the hydrophobic packing of two core helices. As an intact Glu-343/Arg-378 bridge is the default state in unbound ETS1 and maintained in high-affinity and nonspecific complexes, the low-affinity complex represents a unique conformational adaptation to the suboptimization of developmental enhancers.
Keywords: DNA binding protein; DNA sequence motif; DNA-binding domain; ETS proto-oncogene 1 transcription factor (ETS1); ETS transcription factor family; allosteric regulation; developmental factor; low-affinity DNA binding; molecular dynamics; suboptimization; transcription enhancer; transcription factor.
© 2019 Huang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

