Clonal evolution patterns in acute myeloid leukemia with NPM1 mutation
- PMID: 31048683
- PMCID: PMC6497712
- DOI: 10.1038/s41467-019-09745-2
Clonal evolution patterns in acute myeloid leukemia with NPM1 mutation
Abstract
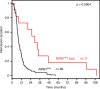
Mutations in the nucleophosmin 1 (NPM1) gene are considered founder mutations in the pathogenesis of acute myeloid leukemia (AML). To characterize the genetic composition of NPM1 mutated (NPM1mut) AML, we assess mutation status of five recurrently mutated oncogenes in 129 paired NPM1mut samples obtained at diagnosis and relapse. We find a substantial shift in the genetic pattern from diagnosis to relapse including NPM1mut loss (n = 11). To better understand these NPM1mut loss cases, we perform whole exome sequencing (WES) and RNA-Seq. At the time of relapse, NPM1mut loss patients (pts) feature distinct mutational patterns that share almost no somatic mutation with the corresponding diagnosis sample and impact different signaling pathways. In contrast, profiles of pts with persistent NPM1mut are reflected by a high overlap of mutations between diagnosis and relapse. Our findings confirm that relapse often originates from persistent leukemic clones, though NPM1mut loss cases suggest a second "de novo" or treatment-associated AML (tAML) as alternative cause of relapse.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

