Three phylogenetic groups have driven the recent population expansion of Cryptococcus neoformans
- PMID: 31048698
- PMCID: PMC6497710
- DOI: 10.1038/s41467-019-10092-5
Three phylogenetic groups have driven the recent population expansion of Cryptococcus neoformans
Abstract
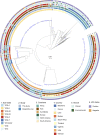
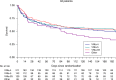
Cryptococcus neoformans (C. neoformans var. grubii) is an environmentally acquired pathogen causing 181,000 HIV-associated deaths each year. We sequenced 699 isolates, primarily C. neoformans from HIV-infected patients, from 5 countries in Asia and Africa. The phylogeny of C. neoformans reveals a recent exponential population expansion, consistent with the increase in the number of susceptible hosts. In our study population, this expansion has been driven by three sub-clades of the C. neoformans VNIa lineage; VNIa-4, VNIa-5 and VNIa-93. These three sub-clades account for 91% of clinical isolates sequenced in our study. Combining the genome data with clinical information, we find that the VNIa-93 sub-clade, the most common sub-clade in Uganda and Malawi, was associated with better outcomes than VNIa-4 and VNIa-5, which predominate in Southeast Asia. This study lays the foundation for further work investigating the dominance of VNIa-4, VNIa-5 and VNIa-93 and the association between lineage and clinical phenotype.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

