Variants in myelin regulatory factor (MYRF) cause autosomal dominant and syndromic nanophthalmos in humans and retinal degeneration in mice
- PMID: 31048900
- PMCID: PMC6527243
- DOI: 10.1371/journal.pgen.1008130
Variants in myelin regulatory factor (MYRF) cause autosomal dominant and syndromic nanophthalmos in humans and retinal degeneration in mice
Abstract
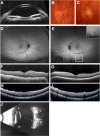
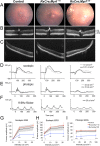
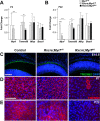
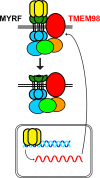
Nanophthalmos is a rare, potentially devastating eye condition characterized by small eyes with relatively normal anatomy, a high hyperopic refractive error, and frequent association with angle closure glaucoma and vision loss. The condition constitutes the extreme of hyperopia or farsightedness, a common refractive error that is associated with strabismus and amblyopia in children. NNO1 was the first mapped nanophthalmos locus. We used combined pooled exome sequencing and strong linkage data in the large family used to map this locus to identify a canonical splice site alteration upstream of the last exon of the gene encoding myelin regulatory factor (MYRF c.3376-1G>A), a membrane bound transcription factor that undergoes autoproteolytic cleavage for nuclear localization. This variant produced a stable RNA transcript, leading to a frameshift mutation p.Gly1126Valfs*31 in the C-terminus of the protein. In addition, we identified an early truncating MYRF frameshift mutation, c.769dupC (p.S264QfsX74), in a patient with extreme axial hyperopia and syndromic features. Myrf conditional knockout mice (CKO) developed depigmentation of the retinal pigment epithelium (RPE) and retinal degeneration supporting a role of this gene in retinal and RPE development. Furthermore, we demonstrated the reduced expression of Tmem98, another known nanophthalmos gene, in Myrf CKO mice, and the physical interaction of MYRF with TMEM98. Our study establishes MYRF as a nanophthalmos gene and uncovers a new pathway for eye growth and development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Autosomal dominant nanophthalmos and high hyperopia associated with a C-terminal frameshift variant in MYRF.Mol Vis. 2019 Sep 21;25:527-534. eCollection 2019. Mol Vis. 2019. PMID: 31700225 Free PMC article.
-
Novel TMEM98 mutations in pedigrees with autosomal dominant nanophthalmos.Mol Vis. 2015 Sep 1;21:1017-23. eCollection 2015. Mol Vis. 2015. PMID: 26392740 Free PMC article.
-
The genetic and clinical landscape of nanophthalmos and posterior microphthalmos in an Australian cohort.Clin Genet. 2020 May;97(5):764-769. doi: 10.1111/cge.13722. Epub 2020 Mar 5. Clin Genet. 2020. PMID: 32052405 Free PMC article.
-
Clinical update in nanophthalmos: Features, diseases and complications associated.Arch Soc Esp Oftalmol (Engl Ed). 2023 Dec;98(12):687-702. doi: 10.1016/j.oftale.2023.10.003. Epub 2023 Oct 7. Arch Soc Esp Oftalmol (Engl Ed). 2023. PMID: 37813187 Review.
-
[Glaucoma and nanophthalmos].Ophthalmologe. 2019 May;116(5):415-422. doi: 10.1007/s00347-018-0835-5. Ophthalmologe. 2019. PMID: 30552473 Review. German.
Cited by
-
Morphological features of anterior segment: factors influencing intraocular pressure after cataract surgery in nanophthalmos.Eye Vis (Lond). 2020 Sep 9;7:47. doi: 10.1186/s40662-020-00212-4. eCollection 2020. Eye Vis (Lond). 2020. PMID: 32974413 Free PMC article.
-
Autosomal dominant nanophthalmos and high hyperopia associated with a C-terminal frameshift variant in MYRF.Mol Vis. 2019 Sep 21;25:527-534. eCollection 2019. Mol Vis. 2019. PMID: 31700225 Free PMC article.
-
Myelin regulatory factor (MYRF) is a critical early regulator of retinal pigment epithelial development.PLoS Genet. 2025 Apr 15;21(4):e1011670. doi: 10.1371/journal.pgen.1011670. eCollection 2025 Apr. PLoS Genet. 2025. PMID: 40233131 Free PMC article.
-
A Glance at the Molecules That Regulate Oligodendrocyte Myelination.Curr Issues Mol Biol. 2022 May 15;44(5):2194-2216. doi: 10.3390/cimb44050149. Curr Issues Mol Biol. 2022. PMID: 35678678 Free PMC article. Review.
-
MYRF: A New Regulator of Cardiac and Early Gonadal Development-Insights from Single Cell RNA Sequencing Analysis.J Clin Med. 2022 Aug 18;11(16):4858. doi: 10.3390/jcm11164858. J Clin Med. 2022. PMID: 36013096 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

