Germline-focussed analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group
- PMID: 31050713
- PMCID: PMC6683854
- DOI: 10.1093/annonc/mdz136
Germline-focussed analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group
Erratum in
-
Erratum to 'Germline-focussed analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group': [Annals of Oncology 30 (2019) 1221-1231].Ann Oncol. 2021 Aug;32(8):1069-1071. doi: 10.1016/j.annonc.2021.05.798. Epub 2021 Jun 2. Ann Oncol. 2021. PMID: 34090768 Free PMC article. No abstract available.
Abstract
It is increasingly common in oncology practice to perform tumour sequencing using large cancer panels. For pathogenic sequence variants in cancer susceptibility genes identified on tumour-only sequencing, it is often unclear whether they are of somatic or constitutional (germline) origin. There is wide-spread disparity regarding both the extent to which systematic 'germline-focussed analysis' is carried out upon tumour sequencing data and for which variants follow-up analysis of a germline sample is carried out. Here we present analyses of paired sequencing data from 17 152 cancer samples, in which 1494 pathogenic sequence variants were identified across 65 cancer susceptibility genes. From these analyses, the European Society of Medical Oncology Precision Medicine Working Group Germline Subgroup has generated (i) recommendations regarding germline-focussed analyses of tumour-only sequencing data, (ii) indications for germline follow-up testing and (iii) guidance on patient information-giving and consent.
Keywords: gene; germline; panel; predisposition; sequencing; susceptibility.
© The Author(s) 2019. Published by Oxford University Press on behalf of the European Society for Medical Oncology.
Figures
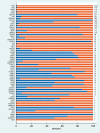
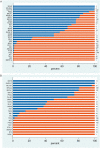


References
-
- Garcia EP, Minkovsky A, Jia Y. et al. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med 2017; 141(6): 751–758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

