Trans Effects on Gene Expression Can Drive Omnigenic Inheritance
- PMID: 31051098
- PMCID: PMC6553491
- DOI: 10.1016/j.cell.2019.04.014
Trans Effects on Gene Expression Can Drive Omnigenic Inheritance
Abstract
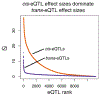
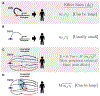
Early genome-wide association studies (GWASs) led to the surprising discovery that, for typical complex traits, most of the heritability is due to huge numbers of common variants with tiny effect sizes. Previously, we argued that new models are needed to understand these patterns. Here, we provide a formal model in which genetic contributions to complex traits are partitioned into direct effects from core genes and indirect effects from peripheral genes acting in trans. We propose that most heritability is driven by weak trans-eQTL SNPs, whose effects are mediated through peripheral genes to impact the expression of core genes. In particular, if the core genes for a trait tend to be co-regulated, then the effects of peripheral variation can be amplified such that nearly all of the genetic variance is driven by weak trans effects. Thus, our model proposes a framework for understanding key features of the architecture of complex traits.
Keywords: cis-eQTLs; complex traits; core genes; genetic architecture; heritability; omnigenic model; polygenic model; trans-eQTLs.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors have no conflict of interest to declare.
Figures
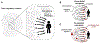
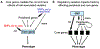
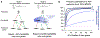
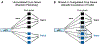
References
-
- Barton NH, Etheridge AM, and Vèber A (2017). The infinitesimal model: Definition, derivation, and implications. Theor. Popul. Biol. 118, 50–73. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

