Viral Metagenomics on Cerebrospinal Fluid
- PMID: 31052348
- PMCID: PMC6562652
- DOI: 10.3390/genes10050332
Viral Metagenomics on Cerebrospinal Fluid
Abstract
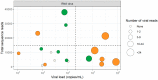
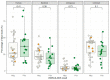
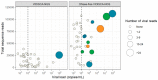
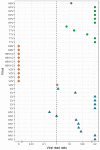
Identifying the causative pathogen in central nervous system (CNS) infections is crucial for patient management and prognosis. Many viruses can cause CNS infections, yet screening for each individually is costly and time-consuming. Most metagenomic assays can theoretically detect all pathogens, but often fail to detect viruses because of their small genome and low viral load. Viral metagenomics overcomes this by enrichment of the viral genomic content in a sample. VIDISCA-NGS is one of the available workflows for viral metagenomics, which requires only a small input volume and allows multiplexing of multiple samples per run. The performance of VIDISCA-NGS was tested on 45 cerebrospinal fluid (CSF) samples from patients with suspected CNS infections in which a virus was identified and quantified by polymerase chain reaction. Eighteen were positive for an RNA virus, and 34 for a herpesvirus. VIDISCA-NGS detected all RNA viruses with a viral load >2 × 104 RNA copies/mL (n = 6) and 8 of 12 of the remaining low load samples. Only one herpesvirus was identified by VIDISCA-NGS, however, when withholding a DNase treatment, 11 of 18 samples with a herpesvirus load >104 DNA copies/mL were detected. Our results indicate that VIDISCA-NGS has the capacity to detect low load RNA viruses in CSF. Herpesvirus DNA in clinical samples is probably non-encapsidated and therefore difficult to detect by VIDISCA-NGS.
Keywords: CNS infection; VIDISCA-NGS; cerebrospinal fluid; encephalitis; metagenomics; metaviromics; next-generation sequencing; viromics; virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

