The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria
- PMID: 31052562
- PMCID: PMC6539912
- DOI: 10.3390/molecules24091711
The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria
Abstract
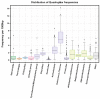
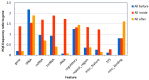
The role of local DNA structures in the regulation of basic cellular processes is an emerging field of research. Amongst local non-B DNA structures, the significance of G-quadruplexes was demonstrated in the last decade, and their presence and functional relevance has been demonstrated in many genomes, including humans. In this study, we analyzed the presence and locations of G-quadruplex-forming sequences by G4Hunter in all complete bacterial genomes available in the NCBI database. G-quadruplex-forming sequences were identified in all species, however the frequency differed significantly across evolutionary groups. The highest frequency of G-quadruplex forming sequences was detected in the subgroup Deinococcus-Thermus, and the lowest frequency in Thermotogae. G-quadruplex forming sequences are non-randomly distributed and are favored in various evolutionary groups. G-quadruplex-forming sequences are enriched in ncRNA segments followed by mRNAs. Analyses of surrounding sequences showed G-quadruplex-forming sequences around tRNA and regulatory sequences. These data point to the unique and non-random localization of G-quadruplex-forming sequences in bacterial genomes.
Keywords: G-quadruplex; G4Hunter; bacteria; bioinformatics; deinococcus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
G-Quadruplexes in the Archaea Domain.Biomolecules. 2020 Sep 21;10(9):1349. doi: 10.3390/biom10091349. Biomolecules. 2020. PMID: 32967357 Free PMC article.
-
Analyses of viral genomes for G-quadruplex forming sequences reveal their correlation with the type of infection.Biochimie. 2021 Jul;186:13-27. doi: 10.1016/j.biochi.2021.03.017. Epub 2021 Apr 9. Biochimie. 2021. PMID: 33839192
-
Re-evaluation of G-quadruplex propensity with G4Hunter.Nucleic Acids Res. 2016 Feb 29;44(4):1746-59. doi: 10.1093/nar/gkw006. Epub 2016 Jan 20. Nucleic Acids Res. 2016. PMID: 26792894 Free PMC article.
-
Biological aspects of DNA/RNA quadruplexes.Biopolymers. 2000-2001;56(3):209-27. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. Biopolymers. 2000. PMID: 11745112 Review.
-
Emerging Role of G-quadruplex DNA as Target in Anticancer Therapy.Curr Pharm Des. 2016;22(44):6612-6624. doi: 10.2174/1381612822666160831101031. Curr Pharm Des. 2016. PMID: 27587203 Review.
Cited by
-
Beyond the Primary Structure of Nucleic Acids: Potential Roles of Epigenetics and Noncanonical Structures in the Regulations of Plant Growth and Stress Responses.Methods Mol Biol. 2023;2642:331-361. doi: 10.1007/978-1-0716-3044-0_18. Methods Mol Biol. 2023. PMID: 36944887
-
Structures and stability of simple DNA repeats from bacteria.Biochem J. 2020 Jan 31;477(2):325-339. doi: 10.1042/BCJ20190703. Biochem J. 2020. PMID: 31967649 Free PMC article. Review.
-
Recurrent Potential G-Quadruplex Sequences in Archaeal Genomes.Front Microbiol. 2021 Mar 24;12:647851. doi: 10.3389/fmicb.2021.647851. eCollection 2021. Front Microbiol. 2021. PMID: 33868206 Free PMC article.
-
Abundance of G-Quadruplex Forming Sequences in the Hepatitis Delta Virus Genomes.ACS Omega. 2024 Jan 9;9(3):4096-4101. doi: 10.1021/acsomega.3c09288. eCollection 2024 Jan 23. ACS Omega. 2024. PMID: 38284014 Free PMC article.
-
The Rich World of p53 DNA Binding Targets: The Role of DNA Structure.Int J Mol Sci. 2019 Nov 9;20(22):5605. doi: 10.3390/ijms20225605. Int J Mol Sci. 2019. PMID: 31717504 Free PMC article. Review.
References
-
- Nelson L.D., Bender C., Mannsperger H., Buergy D., Kambakamba P., Mudduluru G., Korf U., Hughes D., Van Dyke M.W., Allgayer H. Triplex DNA-binding proteins are associated with clinical outcomes revealed by proteomic measurements in patients with colorectal cancer. Mol. Cancer. 2012;11:38. doi: 10.1186/1476-4598-11-38. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 18-15548S/Grant Agency of the Czech Republic
- LO1208 TEWEP/Ministry of Education, Youth and Sports of the Czech Republic in the "National Feasibility Program I"
- CZ.1.05/2.1.00/19.0388/EU structural funding Operational Programme Research and Development for innovation
- SGS/09/PrF/2019/University of Ostrava
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

