The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria
- PMID: 31052562
- PMCID: PMC6539912
- DOI: 10.3390/molecules24091711
The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria
Abstract
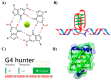
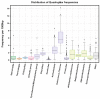
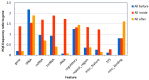
The role of local DNA structures in the regulation of basic cellular processes is an emerging field of research. Amongst local non-B DNA structures, the significance of G-quadruplexes was demonstrated in the last decade, and their presence and functional relevance has been demonstrated in many genomes, including humans. In this study, we analyzed the presence and locations of G-quadruplex-forming sequences by G4Hunter in all complete bacterial genomes available in the NCBI database. G-quadruplex-forming sequences were identified in all species, however the frequency differed significantly across evolutionary groups. The highest frequency of G-quadruplex forming sequences was detected in the subgroup Deinococcus-Thermus, and the lowest frequency in Thermotogae. G-quadruplex forming sequences are non-randomly distributed and are favored in various evolutionary groups. G-quadruplex-forming sequences are enriched in ncRNA segments followed by mRNAs. Analyses of surrounding sequences showed G-quadruplex-forming sequences around tRNA and regulatory sequences. These data point to the unique and non-random localization of G-quadruplex-forming sequences in bacterial genomes.
Keywords: G-quadruplex; G4Hunter; bacteria; bioinformatics; deinococcus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Nelson L.D., Bender C., Mannsperger H., Buergy D., Kambakamba P., Mudduluru G., Korf U., Hughes D., Van Dyke M.W., Allgayer H. Triplex DNA-binding proteins are associated with clinical outcomes revealed by proteomic measurements in patients with colorectal cancer. Mol. Cancer. 2012;11:38. doi: 10.1186/1476-4598-11-38. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 18-15548S/Grant Agency of the Czech Republic
- LO1208 TEWEP/Ministry of Education, Youth and Sports of the Czech Republic in the "National Feasibility Program I"
- CZ.1.05/2.1.00/19.0388/EU structural funding Operational Programme Research and Development for innovation
- SGS/09/PrF/2019/University of Ostrava
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

