Comparing sequence and structure of falcipains and human homologs at prodomain and catalytic active site for malarial peptide based inhibitor design
- PMID: 31053072
- PMCID: PMC6500056
- DOI: 10.1186/s12936-019-2790-2
Comparing sequence and structure of falcipains and human homologs at prodomain and catalytic active site for malarial peptide based inhibitor design
Abstract
Background: Falcipains are major cysteine proteases of Plasmodium falciparum involved in haemoglobin degradation and remain attractive anti-malarial drug targets. Several inhibitors against these proteases have been identified, yet none of them has been approved for malaria treatment. Other Plasmodium species also possess highly homologous proteins to falcipains. For selective therapeutic targeting, identification of sequence and structure differences with homologous human cathepsins is necessary. The substrate processing activity of these proteins is tightly controlled via a prodomain segment occluding the active site which is chopped under low pH conditions exposing the catalytic site. Current work characterizes these proteases to identify residues mediating the prodomain regulatory function for the design of peptide based anti-malarial inhibitors.
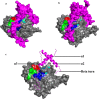
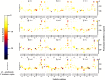
Methods: Sequence and structure variations between prodomain regions of plasmodial proteins and human cathepsins were determined using in silico approaches. Additionally, evolutionary clustering of these proteins was evaluated using phylogenetic analysis. High quality partial zymogen protein structures were modelled using homology modelling and residue interaction analysis performed between the prodomain segment and mature domain to identify key interacting residues between these two domains. The resulting information was used to determine short peptide sequences which could mimic the inherent regulatory function of the prodomain regions. Through flexible docking, the binding affinity of proposed peptides on the proteins studied was evaluated.
Results: Sequence, evolutionary and motif analyses showed important differences between plasmodial and human proteins. Residue interaction analysis identified important residues crucial for maintaining prodomain integrity across the different proteins as well as the pro-segment responsible for inhibitory mechanism. Binding affinity of suggested peptides was highly dependent on their residue composition and length.
Conclusions: Despite the conserved structural and catalytic mechanism between human cathepsins and plasmodial proteases, current work revealed significant differences between the two protein groups which may provide valuable information for selective anti-malarial inhibitor development. Part of this study aimed to design peptide inhibitors based on endogenous inhibitory portions of protease prodomains as a novel aspect. Even though peptide inhibitors may not be practical solutions to malaria at this stage, the approach followed and results offer a promising means to find new malarial inhibitors.
Keywords: Binding affinity; Cysteine protease; Falcipain; Homology modelling; Prodomain inhibitory segment; Zymogen.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures









References
-
- WHO. World malaria report 2017. Geneva: World Health Organization; 2017. http://apps.who.int/iris/bitstream/handle/10665/259492/9789241565523-eng.... Accessed 8 May 2018.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

