Systematic characterization of germline variants from the DiscovEHR study endometrial carcinoma population
- PMID: 31053132
- PMCID: PMC6499978
- DOI: 10.1186/s12920-019-0504-9
Systematic characterization of germline variants from the DiscovEHR study endometrial carcinoma population
Erratum in
-
Correction to: Systematic characterization of germline variants from the DiscovEHR study endometrial carcinoma population.BMC Med Genomics. 2019 May 22;12(1):65. doi: 10.1186/s12920-019-0523-6. BMC Med Genomics. 2019. PMID: 31118041 Free PMC article.
Abstract
Background: Endometrial cancer (EMCA) is the fifth most common cancer among women in the world. Identification of potentially pathogenic germline variants from individuals with EMCA will help characterize genetic features that underlie the disease and potentially predispose individuals to its pathogenesis.
Methods: The Geisinger Health System's (GHS) DiscovEHR cohort includes exome sequencing on over 50,000 consenting patients, 297 of whom have evidence of an EMCA diagnosis in their electronic health record. Here, rare variants were annotated as potentially pathogenic.
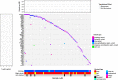
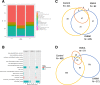
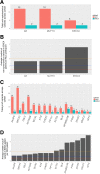
Results: Eight genes were identified as having increased burden in the EMCA cohort relative to the non-cancer control cohort. None of the eight genes had an increased burden in the other hormone related cancer cohort from GHS, suggesting they can help characterize the underlying genetic variation that gives rise to EMCA. Comparing GHS to the cancer genome atlas (TCGA) EMCA germline data illustrated 34 genes with potentially pathogenic variation and eight unique potentially pathogenic variants that were present in both studies. Thus, similar germline variation among genes can be observed in unique EMCA cohorts and could help prioritize genes to investigate for future work.
Conclusion: In summary, this systematic characterization of potentially pathogenic germline variants describes the genetic underpinnings of EMCA through the use of data from a single hospital system.
Keywords: DiscovEHR; Endometrial Cancer; Germline variants; TCGA; Uterine Cancer; Whole exome sequencing.
Conflict of interest statement
Ethics approval and consent to participate
The DiscovEHR cohort enrolled individuals from MyCode, which uses written consent and has previously been described [30]. This study was deemed to be exempt, therefore no further consent was needed. Geisinger Medical Ethics Committee – Geisinger Health System IRB protocol. “Phenotype validation and Biomarker Discovery Protocol for gynecological disorders”. IRB number 2016–0119 was approved on 03/10/2016.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Braun MM, Overbeek-Wager EA, Grumbo RJ. Diagnosis and Management of Endometrial Cancer. Am Fam Physician. 2016;93:468–474. - PubMed
-
- Hoskins WJ. Principles and practice of gynecologic oncology: Lippincott Williams & Wilkins; 2005. https://books.google.com/books?hl=en&lr=&id=KW9esgo759EC&oi=fnd&pg=PR11&....
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

