Structure and Dynamics of the Central Lipid Pool and Proteins of the Bacterial Holo-Translocon
- PMID: 31053257
- PMCID: PMC6531790
- DOI: 10.1016/j.bpj.2019.04.002
Structure and Dynamics of the Central Lipid Pool and Proteins of the Bacterial Holo-Translocon
Abstract
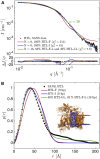
The bacterial Sec translocon, SecYEG, associates with accessory proteins YidC and the SecDF-YajC subcomplex to form the bacterial holo-translocon (HTL). The HTL is a dynamic and flexible protein transport machine capable of coordinating protein secretion across the membrane and efficient lateral insertion of nascent membrane proteins. It has been hypothesized that a central lipid core facilitates the controlled passage of membrane proteins into the bilayer, ensuring the efficient formation of their native state. By performing small-angle neutron scattering on protein solubilized in "match-out" deuterated detergent, we have been able to interrogate a "naked" HTL complex, with the scattering contribution of the surrounding detergent micelle rendered invisible. Such an approach has allowed the confirmation of a lipid core within the HTL, which accommodates between 8 and 29 lipids. Coarse-grained molecular dynamics simulations of the HTL also demonstrate a dynamic, central pool of lipids. An opening at this lipid-rich region between YidC and the SecY lateral gate may provide an exit gateway for newly synthesized, correctly oriented, membrane protein helices, or even small bundles of helices, to emerge from the HTL.
Copyright © 2019 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures





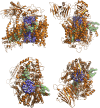
References
-
- Brundage L., Hendrick J.P., Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. - PubMed
-
- Görlich D., Rapoport T.A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. - PubMed
-
- Rapoport T.A., Li L., Park E. Structural and Mechanistic Insights into Protein Translocation. Annu. Rev. Cell Dev. Biol. 2017;33:369–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

